Pre-TCR ligand binding impacts thymocyte development before αβTCR expression
- PMID: 26056289
- PMCID: PMC4500245
- DOI: 10.1073/pnas.1504971112
Pre-TCR ligand binding impacts thymocyte development before αβTCR expression
Abstract
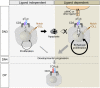
Adaptive cellular immunity requires accurate self- vs. nonself-discrimination to protect against infections and tumorous transformations while at the same time excluding autoimmunity. This vital capability is programmed in the thymus through selection of αβT-cell receptors (αβTCRs) recognizing peptides bound to MHC molecules (pMHC). Here, we show that the pre-TCR (preTCR), a pTα-β heterodimer appearing before αβTCR expression, directs a previously unappreciated initial phase of repertoire selection. Contrasting with the ligand-independent model of preTCR function, we reveal through NMR and bioforce-probe analyses that the β-subunit binds pMHC using Vβ complementarity-determining regions as well as an exposed hydrophobic Vβ patch characteristic of the preTCR. Force-regulated single bonds akin to those of αβTCRs but with more promiscuous ligand specificity trigger calcium flux. Thus, thymic development involves sequential β- and then, αβ-repertoire tuning, whereby preTCR interactions with self pMHC modulate early thymocyte expansion, with implications for β-selection, immunodominant peptide recognition, and germ line-encoded MHC interaction.
Keywords: NMR spectroscopy; biomembrane force probe; pre–T-cell receptor; repertoire selection; thymic development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

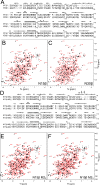

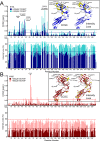

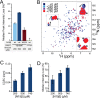
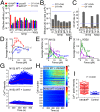
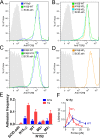
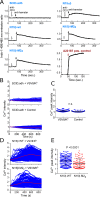
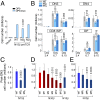


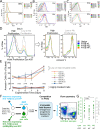
Comment in
-
Pre-T-cell receptor binds MHC: Implications for thymocyte signaling and selection.Proc Natl Acad Sci U S A. 2015 Jul 7;112(27):8166-7. doi: 10.1073/pnas.1510127112. Epub 2015 Jul 1. Proc Natl Acad Sci U S A. 2015. PMID: 26134398 Free PMC article. No abstract available.
References
-
- Prockop S, Petrie HT. Cell migration and the anatomic control of thymocyte precursor differentiation. Semin Immunol. 2000;12(5):435–444. - PubMed
-
- Shah DK, Zúñiga-Pflücker JC. An overview of the intrathymic intricacies of T cell development. J Immunol. 2014;192(9):4017–4023. - PubMed
-
- von Boehmer H. The thymus in immunity and in malignancy. Cancer Immunol Res. 2014;2(7):592–597. - PubMed
-
- Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. - PubMed
-
- Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402(6759):255–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI37581/AI/NIAID NIH HHS/United States
- R01 AI044902/AI/NIAID NIH HHS/United States
- R01 AI037581/AI/NIAID NIH HHS/United States
- P01GM04746/GM/NIGMS NIH HHS/United States
- R01AI088023/AI/NIAID NIH HHS/United States
- P01 GM047467/GM/NIGMS NIH HHS/United States
- R01 AI088023/AI/NIAID NIH HHS/United States
- R01AI100643/AI/NIAID NIH HHS/United States
- R56 AI019807/AI/NIAID NIH HHS/United States
- R01 AI100643/AI/NIAID NIH HHS/United States
- R56 AI088023/AI/NIAID NIH HHS/United States
- R01AI044902/AI/NIAID NIH HHS/United States
- R01AI19807/AI/NIAID NIH HHS/United States
- R21 AI044902/AI/NIAID NIH HHS/United States
- R01 AI019807/AI/NIAID NIH HHS/United States
- R37 AI019807/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

