The chromatin fiber: multiscale problems and approaches
- PMID: 26057099
- PMCID: PMC4476956
- DOI: 10.1016/j.sbi.2015.04.002
The chromatin fiber: multiscale problems and approaches
Abstract
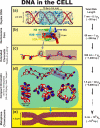
The structure of chromatin, affected by many factors from DNA linker lengths to posttranslational modifications, is crucial to the regulation of eukaryotic cells. Combined experimental and computational methods have led to new insights into its structural and dynamical features, from interactions due to the flexible core histone tails or linker histones to the physical mechanism driving the formation of chromosomal domains. Here we present a perspective of recent advances in chromatin modeling techniques at the atomic, mesoscopic, and chromosomal scales with a view toward developing multiscale computational strategies to integrate such findings. Innovative modeling methods that connect molecular to chromosomal scales are crucial for interpreting experiments and eventually deciphering the complex dynamic organization and function of chromatin in the cell.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


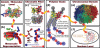
References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Grigoryev S, Arya G, Correll S, Woodcock C, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci USA. 2009;106:13317–13322. [Pioneering cross-linking experiment combined with mesoscale modeling shows that the two different models—zigzag and solenoid fibers—could be merged into one heteromorphic compact fiber with enhanced i±1 internucleosome interactions due to small percentage (Ȉ20%) of linker DNA bending. Such a fiber can be dominant in moderate divalent ion concentrations.] - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

