Ligand-Induced Proton Transfer and Low-Barrier Hydrogen Bond Revealed by X-ray Crystallography
- PMID: 26057252
- PMCID: PMC4530788
- DOI: 10.1021/jacs.5b00749
Ligand-Induced Proton Transfer and Low-Barrier Hydrogen Bond Revealed by X-ray Crystallography
Abstract
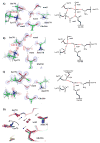
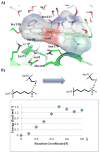
Ligand binding can change the pKa of protein residues and influence enzyme catalysis. Herein, we report three ultrahigh resolution X-ray crystal structures of CTX-M β-lactamase, directly visualizing protonation state changes along the enzymatic pathway: apo protein at 0.79 Å, precovalent complex with nonelectrophilic ligand at 0.89 Å, and acylation transition state (TS) analogue at 0.84 Å. Binding of the noncovalent ligand induces a proton transfer from the catalytic Ser70 to the negatively charged Glu166, and the formation of a low-barrier hydrogen bond (LBHB) between Ser70 and Lys73, with a length of 2.53 Å and the shared hydrogen equidistant from the heteroatoms. QM/MM reaction path calculations determined the proton transfer barrier to be 1.53 kcal/mol. The LBHB is absent in the other two structures although Glu166 remains neutral in the covalent complex. Our data represents the first X-ray crystallographic example of a hydrogen engaged in an enzymatic LBHB, and demonstrates that desolvation of the active site by ligand binding can provide a protein microenvironment conducive to LBHB formation. It also suggests that LBHBs may contribute to stabilization of the TS in general acid/base catalysis together with other preorganized features of enzyme active sites. These structures reconcile previous experimental results suggesting alternatively Glu166 or Lys73 as the general base for acylation, and underline the importance of considering residue protonation state change when modeling protein-ligand interactions. Additionally, the observation of another LBHB (2.47 Å) between two conserved residues, Asp233 and Asp246, suggests that LBHBs may potentially play a special structural role in proteins.
Figures



References
-
- Cleland WW, Kreevoy MM. Science. 1994;264:1887. - PubMed
-
- Halkides CJ, Wu YQ, Murray CJ. Biochemistry. 1996;35:15941. - PubMed
-
- Graham JD, Buytendyk AM, Wang D, Bowen KH, Collins KD. Biochemistry. 2014;53:344. - PubMed
-
- Frey PA, Whitt SA, Tobin JB. Science. 1994;264:1927. - PubMed
-
- Westler WM, Weinhold F, Markley JL. J Am Chem Soc. 2002;124:14373. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions

