A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea
- PMID: 26058368
- PMCID: PMC4461920
- DOI: 10.1038/srep11166
A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea
Abstract
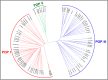
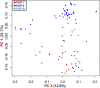
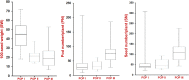
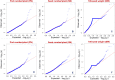
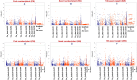
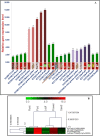
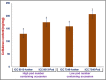
We identified 44844 high-quality SNPs by sequencing 92 diverse chickpea accessions belonging to a seed and pod trait-specific association panel using reference genome- and de novo-based GBS (genotyping-by-sequencing) assays. A GWAS (genome-wide association study) in an association panel of 211, including the 92 sequenced accessions, identified 22 major genomic loci showing significant association (explaining 23-47% phenotypic variation) with pod and seed number/plant and 100-seed weight. Eighteen trait-regulatory major genomic loci underlying 13 robust QTLs were validated and mapped on an intra-specific genetic linkage map by QTL mapping. A combinatorial approach of GWAS, QTL mapping and gene haplotype-specific LD mapping and transcript profiling uncovered one superior haplotype and favourable natural allelic variants in the upstream regulatory region of a CesA-type cellulose synthase (Ca_Kabuli_CesA3) gene regulating high pod and seed number/plant (explaining 47% phenotypic variation) in chickpea. The up-regulation of this superior gene haplotype correlated with increased transcript expression of Ca_Kabuli_CesA3 gene in the pollen and pod of high pod/seed number accession, resulting in higher cellulose accumulation for normal pollen and pollen tube growth. A rapid combinatorial genome-wide SNP genotyping-based approach has potential to dissect complex quantitative agronomic traits and delineate trait-regulatory genomic loci (candidate genes) for genetic enhancement in crop plants, including chickpea.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Cho S. et al. Mapping genes for double podding and other morphological traits in chickpea. Euphytica 125, 285–292 (2002).
-
- Rakshit S. et al. DAF marker tightly linked to a major locus for Ascochyta blight resistance in chickpea (Cicer arietinum L.). Euphytica 132, 23–30 (2003).
-
- Sharma K. D., Winter P., Kahl G. & Muehlbauer F. J. Molecular mapping of Fusarium oxysporum f. sp. ciceris race 3 resistance gene in chickpea. Theor. Appl. Genet. 108, 1243–1248 (2004). - PubMed
-
- Sharma K. D., Chen W. & Muehlbauer F. J. Genetics of chickpea resistance to five races of Fusarium wilt and a concise set of race differentials for Fusarium oxysporum f.sp. ciceris. Plant Dis. 89, 385–390 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

