Genome-Wide Identification of Klebsiella pneumoniae Fitness Genes during Lung Infection
- PMID: 26060277
- PMCID: PMC4462621
- DOI: 10.1128/mBio.00775-15
Genome-Wide Identification of Klebsiella pneumoniae Fitness Genes during Lung Infection
Abstract
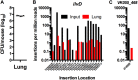
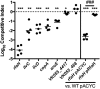
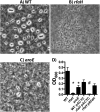
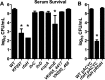
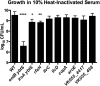
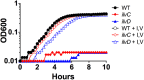
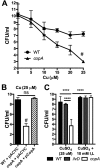
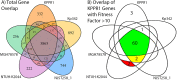
Klebsiella pneumoniae is an urgent public health threat because of resistance to carbapenems, antibiotics of last resort against Gram-negative bacterial infections. Despite the fact that K. pneumoniae is a leading cause of pneumonia in hospitalized patients, the bacterial factors required to cause disease are poorly understood. Insertion site sequencing combines transposon mutagenesis with high-throughput sequencing to simultaneously screen thousands of insertion mutants for fitness defects during infection. Using the recently sequenced K. pneumoniae strain KPPR1 in a well-established mouse model of pneumonia, insertion site sequencing was performed on a pool of >25,000 transposon mutants. The relative fitness requirement of each gene was ranked based on the ratio of lung to inoculum read counts and concordance between insertions in the same gene. This analysis revealed over 300 mutants with at least a 2-fold fitness defect and 69 with defects ranging from 10- to >2,000-fold. Construction of 6 isogenic mutants for use in competitive infections with the wild type confirmed their requirement for lung fitness. Critical fitness genes included those for the synthesis of branched-chain and aromatic amino acids that are essential in mice and humans, the transcriptional elongation factor RfaH, and the copper efflux pump CopA. The majority of fitness genes were conserved among reference strains representative of diverse pathotypes. These results indicate that regulation of outer membrane components and synthesis of amino acids that are essential to its host are critical for K. pneumoniae fitness in the lung.
Importance: Klebsiella pneumoniae is a bacterium that commonly causes pneumonia in patients after they are admitted to the hospital. K. pneumoniae is becoming resistant to all available antibiotics, and when these infections spread to the bloodstream, over half of patients die. Since currently available antibiotics are failing, we must discover new ways to treat these infections. In this study, we asked what genes the bacterium needs to cause an infection, since the proteins encoded by these genes could be targets for new antibiotics. We identified over 300 genes that K. pneumoniae requires to grow in a mouse model of pneumonia. Many of the genes that we identified are found in K. pneumoniae isolates from throughout the world, including antibiotic-resistant forms. If new antibiotics could be made against the proteins that these genes encode, they may be broadly effective against K. pneumoniae.
Copyright © 2015 Bachman et al.
Figures
References
-
- Centers for Disease Control and Prevention 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

