Identification of Owl Monkey CD4 Receptors Broadly Compatible with Early-Stage HIV-1 Isolates
- PMID: 26063421
- PMCID: PMC4524261
- DOI: 10.1128/JVI.00890-15
Identification of Owl Monkey CD4 Receptors Broadly Compatible with Early-Stage HIV-1 Isolates
Abstract
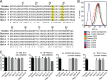
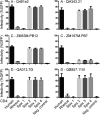
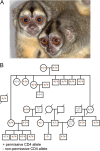
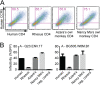
Most HIV-1 variants isolated from early-stage human infections do not use nonhuman primate versions of the CD4 receptor for cellular entry, or they do so poorly. We and others have previously shown that CD4 has experienced strong natural selection over the course of primate speciation, but it is unclear whether this selection has influenced the functional characteristics of CD4 as an HIV-1 receptor. Surprisingly, we find that selection on CD4 has been most intense in the New World monkeys, animals that have never been found to harbor lentiviruses related to HIV-1. Based on this, we sampled CD4 genetic diversity within populations of individuals from seven different species, including five species of New World monkeys. We found that some, but not all, CD4 alleles found in Spix's owl monkeys (Aotus vociferans) encode functional receptors for early-stage human HIV-1 isolates representing all of the major group M clades (A, B, C, and D). However, only some isolates of HIV-1 subtype C can use the CD4 receptor encoded by permissive Spix's owl monkey alleles. We characterized the prevalence of functional CD4 alleles in a colony of captive Spix's owl monkeys and found that 88% of surveyed individuals are homozygous for permissive CD4 alleles, which encode an asparagine at position 39 of the receptor. We found that the CD4 receptors encoded by two other species of owl monkeys (Aotus azarae and Aotus nancymaae) also serve as functional entry receptors for early-stage isolates of HIV-1.
Importance: Nonhuman primates, particularly macaques, are used for preclinical evaluation of HIV-1 vaccine candidates. However, a significant limitation of the macaque model is the fact that most circulating HIV-1 variants cannot use the macaque CD4 receptor to enter cells and have to be adapted to these species. This is particularly true for viral variants from early stages of infection, which represent the most relevant vaccine targets. In this study, we found that some individuals from captive owl monkey populations harbor CD4 alleles that are compatible with a broad collection of HIV-1 isolates, including those isolated from early in infection in highly affected populations and representing diverse subtypes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

