Characterization of Ribosomal Frameshifting in Theiler's Murine Encephalomyelitis Virus
- PMID: 26063423
- PMCID: PMC4524249
- DOI: 10.1128/JVI.01043-15
Characterization of Ribosomal Frameshifting in Theiler's Murine Encephalomyelitis Virus
Abstract
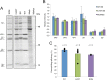
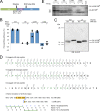
Theiler's murine encephalomyelitis virus (TMEV) is a member of the genus Cardiovirus in the Picornaviridae, a family of positive-sense single-stranded RNA viruses. Previously, we demonstrated that in the related cardiovirus, Encephalomyocarditis virus, a programmed-1 ribosomal frameshift (1 PRF) occurs at a conserved G_GUU_UUU sequence within the 2B-encoding region of the polyprotein open reading frame (ORF). Here we show that-1 PRF occurs at a similar site during translation of the TMEV genome. In addition, we demonstrate that a predicted 3= RNA stem-loop structure at a noncanonical spacing downstream of the shift site is required for efficient frameshifting in TMEV and that frameshifting also requires virus infection. Mutating the G_GUU_UUU shift site to inhibit frameshifting results in an attenuated virus with reduced growth kinetics and a small-plaque phenotype. Frameshifting in the virus context was found to be extremely efficient at 74 to 82%, which, to our knowledge, is the highest frameshifting efficiency recorded to date for any virus. We propose that highly efficient-1 PRF in TMEV provides a mechanism to escape the confines of equimolar expression normally inherent in the single-polyprotein expression strategy of picornaviruses.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

