HJURP is involved in the expansion of centromeric chromatin
- PMID: 26063729
- PMCID: PMC4571335
- DOI: 10.1091/mbc.E15-02-0094
HJURP is involved in the expansion of centromeric chromatin
Abstract
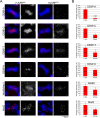
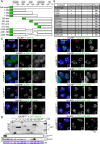
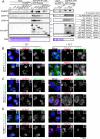
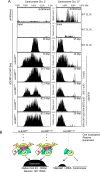
The CENP-A-specific chaperone HJURP mediates CENP-A deposition at centromeres. The N-terminal region of HJURP is responsible for binding to soluble CENP-A. However, it is unclear whether other regions of HJURP have additional functions for centromere formation and maintenance. In this study, we generated chicken DT40 knockout cell lines and gene replacement constructs for HJURP to assess the additional functions of HJURP in vivo. Our analysis revealed that the middle region of HJURP associates with the Mis18 complex protein M18BP1/KNL2 and that the HJURP-M18BP1 association is required for HJURP function. In addition, on the basis of the analysis of artificial centromeres induced by ectopic HJURP localization, we demonstrate that HJURP exhibits a centromere expansion activity that is separable from its CENP-A-binding activity. We also observed centromere expansion surrounding natural centromeres after HJURP overexpression. We propose that this centromere expansion activity reflects the functional properties of HJURP, which uses this activity to contribute to the plastic establishment of a centromeric chromatin structure.
© 2015 Perpelescu et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures


References
-
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. - PubMed
-
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

