YbiB from Escherichia coli, the Defining Member of the Novel TrpD2 Family of Prokaryotic DNA-binding Proteins
- PMID: 26063803
- PMCID: PMC4528117
- DOI: 10.1074/jbc.M114.620575
YbiB from Escherichia coli, the Defining Member of the Novel TrpD2 Family of Prokaryotic DNA-binding Proteins
Abstract
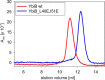
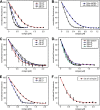
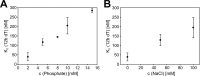
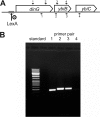
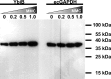
We present the crystal structure and biochemical characterization of Escherichia coli YbiB, a member of the hitherto uncharacterized TrpD2 protein family. Our results demonstrate that the functional diversity of proteins with a common fold can be far greater than predictable by computational annotation. The TrpD2 proteins show high structural homology to anthranilate phosphoribosyltransferase (TrpD) and nucleoside phosphorylase class II enzymes but bind with high affinity (KD = 10-100 nM) to nucleic acids without detectable sequence specificity. The difference in affinity between single- and double-stranded DNA is minor. Results suggest that multiple YbiB molecules bind to one longer DNA molecule in a cooperative manner. The YbiB protein is a homodimer that, therefore, has two electropositive DNA binding grooves. But due to negative cooperativity within the dimer, only one groove binds DNA in in vitro experiments. A monomerized variant remains able to bind DNA with similar affinity, but the negative cooperative effect is eliminated. The ybiB gene forms an operon with the DNA helicase gene dinG and is under LexA control, being induced by DNA-damaging agents. Thus, speculatively, the TrpD2 proteins may be part of the LexA-controlled SOS response in bacteria.
Keywords: DNA damage response; DNA-binding protein; cooperativity; crystallography; dimerization; functional annotation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Innan H., Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108 - PubMed
-
- Marino M., Deuss M., Svergun D. I., Konarev P. V., Sterner R., Mayans O. (2006) Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus. J. Biol. Chem. 281, 21410–21421 - PubMed
-
- Lee C. E., Goodfellow C., Javid-Majd F., Baker E. N., Shaun Lott J. (2006) The crystal structure of TrpD, a metabolic enzyme essential for lung colonization by Mycobacterium tuberculosis, in complex with its substrate phosphoribosylpyrophosphate. J. Mol. Biol. 355, 784–797 - PubMed
-
- Kim C., Xuong N. H., Edwards S., Madhusudan, Yee M. C., Spraggon G., Mills S. E. (2002) The crystal structure of anthranilate phosphoribosyltransferase from the enterobacterium Pectobacterium carotovorum. FEBS Lett. 523, 239–246 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

