TRRUST: a reference database of human transcriptional regulatory interactions
- PMID: 26066708
- PMCID: PMC4464350
- DOI: 10.1038/srep11432
TRRUST: a reference database of human transcriptional regulatory interactions
Abstract
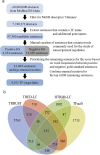
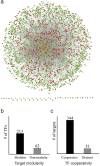
The reconstruction of transcriptional regulatory networks (TRNs) is a long-standing challenge in human genetics. Numerous computational methods have been developed to infer regulatory interactions between human transcriptional factors (TFs) and target genes from high-throughput data, and their performance evaluation requires gold-standard interactions. Here we present a database of literature-curated human TF-target interactions, TRRUST (transcriptional regulatory relationships unravelled by sentence-based text-mining, http://www.grnpedia.org/trrust), which currently contains 8,015 interactions between 748 TF genes and 1,975 non-TF genes. A sentence-based text-mining approach was employed for efficient manual curation of regulatory interactions from approximately 20 million Medline abstracts. To the best of our knowledge, TRRUST is the largest publicly available database of literature-curated human TF-target interactions to date. TRRUST also has several useful features: i) information about the mode-of-regulation; ii) tests for target modularity of a query TF; iii) tests for TF cooperativity of a query target; iv) inferences about cooperating TFs of a query TF; and v) prioritizing associated pathways and diseases with a query TF. We observed high enrichment of TF-target pairs in TRRUST for top-scored interactions inferred from high-throughput data, which suggests that TRRUST provides a reliable benchmark for the computational reconstruction of human TRNs.
Figures


References
-
- Vaquerizas J. M., Kummerfeld S. K., Teichmann S. A. & Luscombe N. M. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10, 252–63 (2009). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

