Molecular genetic mechanisms of allelic specific regulation of murine Comt expression
- PMID: 26067582
- PMCID: PMC4579042
- DOI: 10.1097/j.pain.0000000000000258
Molecular genetic mechanisms of allelic specific regulation of murine Comt expression
Abstract
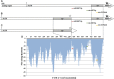
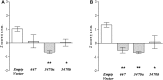
A functional allele of the mouse catechol-O-methyltransferase (Comt) gene is defined by the insertion of a B2 short interspersed repeat element in its 3'-untranslated region (UTR). This allele has been associated with a number of phenotypes, such as pain and anxiety. In comparison with mice carrying the ancestral allele (Comt+), Comt B2i mice show higher Comt mRNA and enzymatic activity levels. Here, we investigated the molecular genetic mechanisms underlying this allelic specific regulation of Comt expression. Insertion of the B2 element introduces an early polyadenylation signal generating a shorter Comt transcript, in addition to the longer ancestral mRNA. Comparative analysis and in silico prediction of Comt mRNA potential targets within the transcript 3' to the B2 element was performed and allowed choosing microRNA (miRNA) candidates for experimental screening: mmu-miR-3470a, mmu-miR-3470b, and mmu-miR-667. Cell transfection with each miRNA downregulated the expression of the ancestral transcript and COMT enzymatic activity. Our in vivo experiments showed that mmu-miR-667-3p is strongly correlated with decreasing amounts of Comt mRNA in the brain, and lentiviral injections of mmu-miR-3470a, mmu-miR-3470b, and mmu-miR-667 increase hypersensitivity in the mouse formalin model, consistent with reduced COMT activity. In summary, our data demonstrate that the Comt+ transcript contains regulatory miRNA signals in its 3'-untranslated region leading to mRNA degradation; these signals, however, are absent in the shorter transcript, resulting in higher mRNA expression and activity levels.
Conflict of interest statement
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

