DNA transposons have colonized the genome of the giant virus Pandoravirus salinus
- PMID: 26067596
- PMCID: PMC4495683
- DOI: 10.1186/s12915-015-0145-1
DNA transposons have colonized the genome of the giant virus Pandoravirus salinus
Abstract
Background: Transposable elements are mobile DNA sequences that are widely distributed in prokaryotic and eukaryotic genomes, where they represent a major force in genome evolution. However, transposable elements have rarely been documented in viruses, and their contribution to viral genome evolution remains largely unexplored. Pandoraviruses are recently described DNA viruses with genome sizes that exceed those of some prokaryotes, rivaling parasitic eukaryotes. These large genomes appear to include substantial noncoding intergenic spaces, which provide potential locations for transposable element insertions. However, no mobile genetic elements have yet been reported in pandoravirus genomes.
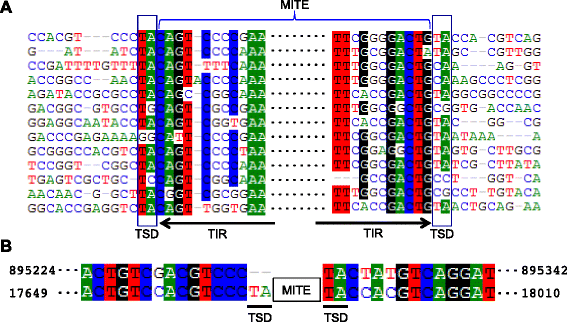
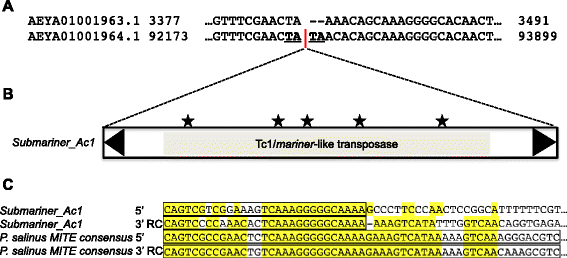
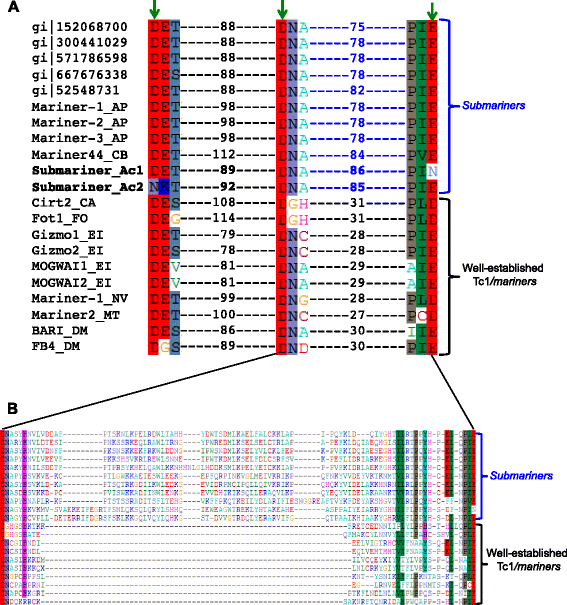
Results: Here, we report a family of miniature inverted-repeat transposable elements (MITEs) in the Pandoravirus salinus genome, representing the first description of a virus populated with a canonical transposable element family that proliferated by transposition within the viral genome. The MITE family, which we name Submariner, includes 30 copies with all the hallmarks of MITEs: short length, terminal inverted repeats, TA target site duplication, and no coding capacity. Submariner elements show signs of transposition and are undetectable in the genome of Pandoravirus dulcis, the closest known relative Pandoravirus salinus. We identified a DNA transposon related to Submariner in the genome of Acanthamoeba castellanii, a species thought to host pandoraviruses, which contains remnants of coding sequence for a Tc1/mariner transposase. These observations suggest that the Submariner MITEs of P. salinus belong to the widespread Tc1/mariner superfamily and may have been mobilized by an amoebozoan host. Ten of the 30 MITEs in the P. salinus genome are located within coding regions of predicted genes, while others are close to genes, suggesting that these transposons may have contributed to viral genetic novelty.
Conclusions: Our discovery highlights the remarkable ability of DNA transposons to colonize and shape genomes from all domains of life, as well as giant viruses. Our findings continue to blur the division between viral and cellular genomes, adhering to the emerging view that the content, dynamics, and evolution of the genomes of giant viruses do not substantially differ from those of cellular organisms.
Figures
References
-
- Craig NL, Craigie R, Gellert M, Mobile LAM, DNA II. Washington. DC: American Society for Microbiology Press; 2002.
-
- Feschotte C, Zhang X, Wessler SR. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: American Society of Microbiology Press; 2002. pp. 1147–58.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

