Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling
- PMID: 26067716
- PMCID: PMC4509974
- DOI: 10.1016/j.tibs.2015.05.001
Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling
Abstract
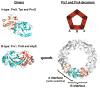
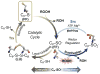
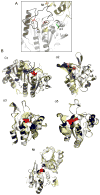
Peroxiredoxins (Prxs) are a ubiquitous family of cysteine-dependent peroxidase enzymes that play dominant roles in regulating peroxide levels within cells. These enzymes, often present at high levels and capable of rapidly clearing peroxides, display a remarkable array of variations in their oligomeric states and susceptibility to regulation by hyperoxidative inactivation and other post-translational modifications. Key conserved residues within the active site promote catalysis by stabilizing the transition state required for transferring the terminal oxygen of hydroperoxides to the active site (peroxidatic) cysteine residue. Extensive investigations continue to expand our understanding of the scope of their importance as well as the structures and forces at play within these critical defense and regulatory enzymes.
Keywords: antioxidant defense; antioxidant enzyme; peroxidase; redox signaling.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


References
-
- Segel IH. Biochemical Calculations. John Wiley & Sons, Inc; 1976.
-
- van der Kamp MW, Mulholland AJ. Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry. 2013;52:2708–2728. - PubMed
-
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

