Dawn and Dusk Set States of the Circadian Oscillator in Sprouting Barley (Hordeum vulgare) Seedlings
- PMID: 26068005
- PMCID: PMC4465908
- DOI: 10.1371/journal.pone.0129781
Dawn and Dusk Set States of the Circadian Oscillator in Sprouting Barley (Hordeum vulgare) Seedlings
Erratum in
-
Correction: Dawn and Dusk Set States of the Circadian Oscillator in Sprouting Barley (Hordeum vulgare) Seedlings.PLoS One. 2015 Sep 11;10(9):e0138255. doi: 10.1371/journal.pone.0138255. eCollection 2015. PLoS One. 2015. PMID: 26359863 Free PMC article. No abstract available.
Abstract
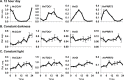
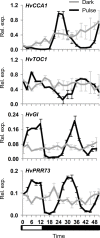
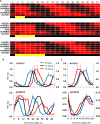
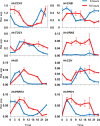
The plant circadian clock is an internal timekeeper that coordinates biological processes with daily changes in the external environment. The transcript levels of clock genes, which oscillate to control circadian outputs, were examined during early seedling development in barley (Hordeum vulgare), a model for temperate cereal crops. Oscillations of clock gene transcript levels do not occur in barley seedlings grown in darkness or constant light but were observed with day-night cycles. A dark-to-light transition influenced transcript levels of some clock genes but triggered only weak oscillations of gene expression, whereas a light-to-dark transition triggered robust oscillations. Single light pulses of 6, 12 or 18 hours induced robust oscillations. The light-to-dark transition was the primary determinant of the timing of subsequent peaks of clock gene expression. After the light-to-dark transition the timing of peak transcript levels of clock gene also varied depending on the length of the preceding light pulse. Thus, a single photoperiod can trigger initiation of photoperiod-dependent circadian rhythms in barley seedlings. Photoperiod-specific rhythms of clock gene expression were observed in two week old barley plants. Changing the timing of dusk altered clock gene expression patterns within a single day, showing that alteration of circadian oscillator behaviour is amongst the most rapid molecular responses to changing photoperiod in barley. A barley EARLY FLOWERING3 mutant, which exhibits rapid photoperiod-insensitive flowering behaviour, does not establish clock rhythms in response to a single photoperiod. The data presented show that dawn and dusk cues are important signals for setting the state of the circadian oscillator during early development of barley and that the circadian oscillator of barley exhibits photoperiod-dependent oscillation states.
Conflict of interest statement
Figures


References
-
- Barak S, Tobin EM, Andronis C, Sugano S, Green RM (2000) All in good time: the Arabidopsis circadian clock. Trends Plant Sci 5: 517–522. - PubMed
-
- McClung CR (2001) Circadian Rhythms in Plants. Ann. Rev. Plant. Biol. 52: 139–162. - PubMed
-
- Millar AJ (2003) Input signals to the plant circadian clock. J Exp Biol 55: 227–283. - PubMed
-
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, et al. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229. - PubMed
-
- Somers DE, Webb AA, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana . Development 125: 485–494. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

