Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels
- PMID: 26068415
- PMCID: PMC4745136
- DOI: 10.1038/ncomms8208
Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels
Abstract
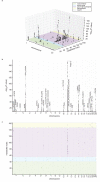
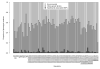
Metabolites are small molecules involved in cellular metabolism, which can be detected in biological samples using metabolomic techniques. Here we present the results of genome-wide association and meta-analyses for variation in the blood serum levels of 129 metabolites as measured by the Biocrates metabolomic platform. In a discovery sample of 7,478 individuals of European descent, we find 4,068 genome- and metabolome-wide significant (Z-test, P < 1.09 × 10(-9)) associations between single-nucleotide polymorphisms (SNPs) and metabolites, involving 59 independent SNPs and 85 metabolites. Five of the fifty-nine independent SNPs are new for serum metabolite levels, and were followed-up for replication in an independent sample (N = 1,182). The novel SNPs are located in or near genes encoding metabolite transporter proteins or enzymes (SLC22A16, ARG1, AGPS and ACSL1) that have demonstrated biomedical or pharmaceutical importance. The further characterization of genetic influences on metabolic phenotypes is important for progress in biological and medical research.
Figures

References
-
- Suhre K, Gieger C. Genetic variation in metabolic phenotypes: study designs and applications. Nat. Rev. Genet. 2012;13:759–769. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

