Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities
- PMID: 26069186
- PMCID: PMC4527963
- DOI: 10.1158/2159-8290.CD-14-1236
Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities
Abstract
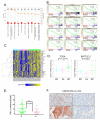
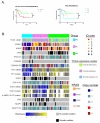
The molecular underpinnings that drive the heterogeneity of KRAS-mutant lung adenocarcinoma are poorly characterized. We performed an integrative analysis of genomic, transcriptomic, and proteomic data from early-stage and chemorefractory lung adenocarcinoma and identified three robust subsets of KRAS-mutant lung adenocarcinoma dominated, respectively, by co-occurring genetic events in STK11/LKB1 (the KL subgroup), TP53 (KP), and CDKN2A/B inactivation coupled with low expression of the NKX2-1 (TTF1) transcription factor (KC). We further revealed biologically and therapeutically relevant differences between the subgroups. KC tumors frequently exhibited mucinous histology and suppressed mTORC1 signaling. KL tumors had high rates of KEAP1 mutational inactivation and expressed lower levels of immune markers, including PD-L1. KP tumors demonstrated higher levels of somatic mutations, inflammatory markers, immune checkpoint effector molecules, and improved relapse-free survival. Differences in drug sensitivity patterns were also observed; notably, KL cells showed increased vulnerability to HSP90-inhibitor therapy. This work provides evidence that co-occurring genomic alterations identify subgroups of KRAS-mutant lung adenocarcinoma with distinct biology and therapeutic vulnerabilities.
Significance: Co-occurring genetic alterations in STK11/LKB1, TP53, and CDKN2A/B-the latter coupled with low TTF1 expression-define three major subgroups of KRAS-mutant lung adenocarcinoma with distinct biology, patterns of immune-system engagement, and therapeutic vulnerabilities.
©2015 American Association for Cancer Research.
Figures





References
-
- Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–81. - PubMed
-
- Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA155196/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA168484/CA/NCI NIH HHS/United States
- 1R01 CA168484-01/CA/NCI NIH HHS/United States
- R01 CA140594/CA/NCI NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- T32 CA009666/CA/NCI NIH HHS/United States
- R01 CA205150/CA/NCI NIH HHS/United States
- P01 CA120964/CA/NCI NIH HHS/United States
- CA016672/CA/NCI NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- 5 P50 CA070907/CA/NCI NIH HHS/United States
- P01 CA154303/CA/NCI NIH HHS/United States
- R01 CA163896/CA/NCI NIH HHS/United States
- R01 CA166480/CA/NCI NIH HHS/United States
- 1 R01 CA155196-01A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

