Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates
- PMID: 26069274
- PMCID: PMC4717365
- DOI: 10.1136/gutjnl-2015-309618
Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates
Abstract
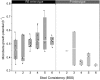
Objective: The assessment of potentially confounding factors affecting colon microbiota composition is essential to the identification of robust microbiome based disease markers. Here, we investigate the link between gut microbiota variation and stool consistency using Bristol Stool Scale classification, which reflects faecal water content and activity, and is considered a proxy for intestinal colon transit time.
Design: Through 16S rDNA Illumina profiling of faecal samples of 53 healthy women, we evaluated associations between microbiome richness, Bacteroidetes:Firmicutes ratio, enterotypes, and genus abundance with self-reported, Bristol Stool Scale-based stool consistency. Each sample's microbiota growth potential was calculated to test whether transit time acts as a selective force on gut bacterial growth rates.
Results: Stool consistency strongly correlates with all known major microbiome markers. It is negatively correlated with species richness, positively associated to the Bacteroidetes:Firmicutes ratio, and linked to Akkermansia and Methanobrevibacter abundance. Enterotypes are distinctly distributed over the BSS-scores. Based on the correlations between microbiota growth potential and stool consistency scores within both enterotypes, we hypothesise that accelerated transit contributes to colon ecosystem differentiation. While shorter transit times can be linked to increased abundance of fast growing species in Ruminococcaceae-Bacteroides samples, hinting to a washout avoidance strategy of faster replication, this trend is absent in Prevotella-enterotyped individuals. Within this enterotype adherence to host tissue therefore appears to be a more likely bacterial strategy to cope with washout.
Conclusions: The strength of the associations between stool consistency and species richness, enterotypes and community composition emphasises the crucial importance of stool consistency assessment in gut metagenome-wide association studies.
Keywords: INTESTINAL BACTERIA; INTESTINAL MICROBIOLOGY.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/
Figures

Comment in
-
Stool consistency as a major confounding factor affecting microbiota composition: an ignored variable?Gut. 2016 Jan;65(1):1-2. doi: 10.1136/gutjnl-2015-310043. Epub 2015 Jul 17. Gut. 2016. PMID: 26187505 No abstract available.
-
Gut microbiota composition associated with stool consistency.Gut. 2016 Mar;65(3):540-2. doi: 10.1136/gutjnl-2015-310328. Epub 2015 Aug 14. Gut. 2016. PMID: 26276682 No abstract available.
-
Stool frequency is associated with gut microbiota composition.Gut. 2017 Mar;66(3):559-560. doi: 10.1136/gutjnl-2016-311935. Epub 2016 Apr 28. Gut. 2017. PMID: 27196592 No abstract available.
-
Water activity does not shape the microbiota in the human colon.Gut. 2017 Oct;66(10):1865-1866. doi: 10.1136/gutjnl-2016-313530. Epub 2017 Jan 6. Gut. 2017. PMID: 28062523 Free PMC article. No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
