Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia
- PMID: 26069293
- PMCID: PMC4800704
- DOI: 10.3324/haematol.2015.124321
Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia
Abstract
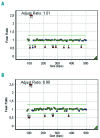
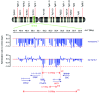
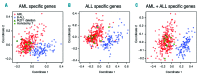
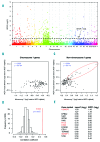
IKAROS family zinc finger 1/IKZF1 is a transcription factor important in lymphoid differentiation, and a known tumor suppressor in acute lymphoid leukemia. Recent studies suggest that IKZF1 is also involved in myeloid differentiation. To investigate whether IKZF1 deletions also play a role in pediatric acute myeloid leukemia, we screened a panel of pediatric acute myeloid leukemia samples for deletions of the IKZF1 locus using multiplex ligation-dependent probe amplification and for mutations using direct sequencing. Three patients were identified with a single amino acid variant without change of IKZF1 length. No frame-shift mutations were found. Out of 11 patients with an IKZF1 deletion, 8 samples revealed a complete loss of chromosome 7, and 3 cases a focal deletion of 0.1-0.9Mb. These deletions included the complete IKZF1 gene (n=2) or exons 1-4 (n=1), all leading to a loss of IKZF1 function. Interestingly, differentially expressed genes in monosomy 7 cases (n=8) when compared to non-deleted samples (n=247) significantly correlated with gene expression changes in focal IKZF1-deleted cases (n=3). Genes with increased expression included genes involved in myeloid cell self-renewal and cell cycle, and a significant portion of GATA target genes and GATA factors. Together, these results suggest that loss of IKZF1 is recurrent in pediatric acute myeloid leukemia and might be a determinant of oncogenesis in acute myeloid leukemia with monosomy 7.
Copyright© Ferrata Storti Foundation.
Figures


References
-
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. - PubMed
-
- Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120(16):3187–3205. - PubMed
-
- Kaspers GJ, Zwaan CM. Pediatric acute myeloid leukemia: towards high-quality cure of all patients. Haematologica. 2007;92(11):1519–1532. - PubMed
-
- Oestreich KJ, Weinmann AS. Ikaros changes the face of NuRD remodeling. Nat Immunol. 2012;13(1):16–18. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

