Structure of potassium channels
- PMID: 26070303
- PMCID: PMC4565861
- DOI: 10.1007/s00018-015-1948-5
Structure of potassium channels
Abstract
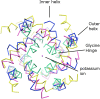
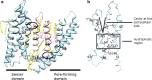
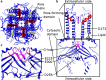
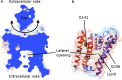
Potassium channels ubiquitously exist in nearly all kingdoms of life and perform diverse but important functions. Since the first atomic structure of a prokaryotic potassium channel (KcsA, a channel from Streptomyces lividans) was determined, tremendous progress has been made in understanding the mechanism of potassium channels and channels conducting other ions. In this review, we discuss the structure of various kinds of potassium channels, including the potassium channel with the pore-forming domain only (KcsA), voltage-gated, inwardly rectifying, tandem pore domain, and ligand-gated ones. The general properties shared by all potassium channels are introduced first, followed by specific features in each class. Our purpose is to help readers to grasp the basic concepts, to be familiar with the property of the different domains, and to understand the structure and function of the potassium channels better.
Keywords: Conductivity; Gating; RCK; Selectivity; Sensor domain.
Conflict of interest statement
No conflict of interest is declared.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

