Making a chocolate chip: development and evaluation of a 6K SNP array for Theobroma cacao
- PMID: 26070980
- PMCID: PMC4535616
- DOI: 10.1093/dnares/dsv009
Making a chocolate chip: development and evaluation of a 6K SNP array for Theobroma cacao
Abstract
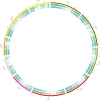
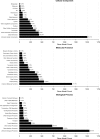
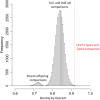
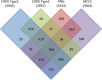
Theobroma cacao, the key ingredient in chocolate production, is one of the world's most important tree fruit crops, with ∼4,000,000 metric tons produced across 50 countries. To move towards gene discovery and marker-assisted breeding in cacao, a single-nucleotide polymorphism (SNP) identification project was undertaken using RNAseq data from 16 diverse cacao cultivars. RNA sequences were aligned to the assembled transcriptome of the cultivar Matina 1-6, and 330,000 SNPs within coding regions were identified. From these SNPs, a subset of 6,000 high-quality SNPs were selected for inclusion on an Illumina Infinium SNP array: the Cacao6kSNP array. Using Cacao6KSNP array data from over 1,000 cacao samples, we demonstrate that our custom array produces a saturated genetic map and can be used to distinguish among even closely related genotypes. Our study enhances and expands the genetic resources available to the cacao research community, and provides the genome-scale set of tools that are critical for advancing breeding with molecular markers in an agricultural species with high genetic diversity.
Keywords: SNP; breeding; cacao; mapping; markers.
© The Author 2015. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures




References
-
- Food and Agriculture Organization of the United Nations. 2015. FAOSTAT. Crops (National Production). http://data.fao.org/ref/29920434-c74e-4ea2-beed-01b832e60609.html?versio... (6 March 2014, date last accessed).
-
- Cuatrecasas J. 1964, Cacao and its allies: a taxonomic revision og the genus Theobroma, Contrib. US Herbarium, 35, 542–3.
-
- Hunter J.R. 1990, The status of cacao (Theobroma-Cacao, Sterculiaceae) in the western-hemisphere, Econ. Bot., 44, 425–39.
-
- Motamayor J.C., Risterucci A.M., Lopez P.A., Ortiz C.F., Moreno A., Lanaud C. 2002, Cacao domestication I: the origin of the cacao cultivated by the Mayas, Heredity, 89, 380–6. - PubMed
-
- Fredholm A. 1911, Breeding of cacao, The West India Committee Circular, 102–3.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

