Regulation and function of the human HSP90AA1 gene
- PMID: 26071189
- PMCID: PMC4519370
- DOI: 10.1016/j.gene.2015.06.018
Regulation and function of the human HSP90AA1 gene
Abstract
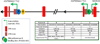
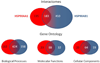
Heat shock protein 90α (Hsp90α), encoded by the HSP90AA1 gene, is the stress inducible isoform of the molecular chaperone Hsp90. Hsp90α is regulated differently and has different functions when compared to the constitutively expressed Hsp90β isoform, despite high amino acid sequence identity between the two proteins. These differences are likely due to variations in nucleotide sequence within non-coding regions, which allows for specific regulation through interaction with particular transcription factors, and to subtle changes in amino acid sequence that allow for unique post-translational modifications. This article will specifically focus on the expression, function and regulation of Hsp90α.
Keywords: Cancer; Drug target; Gene ontology; Gene promoter; Heat shock element; Heat shock protein; Interactome; Molecular chaperone; Post-translational modification; Stress response.
Published by Elsevier B.V.
Figures





References
-
- Ammirante M, Rosati A, Gentilella A, Festa M, Petrella A, Marzullo L, Pascale M, Belisario MA, Leone A, Turco MC. The activity of hsp90α promoter is regulated by NF-κB transcription factors. Oncogene. 2008;27:1175–1178. - PubMed
-
- Banerji U, O’Donnell A, Scurr M, Pacey S, Stapleton S, Asad Y, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. Journal of clinical oncology. American Society of Clinical Oncology. 2005;23(18):4152–4161. - PubMed
-
- Banerji U, Walton M, Raynaud F, Grimshaw R, Kelland L, Valenti M, et al. Pharmacokinetic-pharmacodynamic relationships for the heat shock protein 90 molecular chaperone inhibitor 17-allylamino, 17-demethoxygeldanamycin in human ovarian cancer xenograft models. Clinical cancer research. 2005;11(19 Pt 1):7023–7032. - PubMed
-
- Beebe K, Mollapour M, Scroggins B, Prodromou C, Xu W, Tokita M, Taldone T, Pullen L, Zierer BK, Lee MJ, Trepel J, Buchner J, Bolon D, Chiosis G, Neckers L. Posttranslational modification and conformational state of heat shock protein 90 differentially affect binding of chemically diverse small molecule inhibitors. Oncotarget. 2013;4(7):1065–1074. - PMC - PubMed
-
- Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med. Res. Rev. 2006 May;26(3):310–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

