Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link
- PMID: 26071591
- PMCID: PMC4547899
- DOI: 10.15252/embj.201490878
Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link
Abstract
DNA interstrand cross-links (ICLs) are repaired in S phase by a complex, multistep mechanism involving translesion DNA polymerases. After replication forks collide with an ICL, the leading strand approaches to within one nucleotide of the ICL ("approach"), a nucleotide is inserted across from the unhooked lesion ("insertion"), and the leading strand is extended beyond the lesion ("extension"). How DNA polymerases bypass the ICL is incompletely understood. Here, we use repair of a site-specific ICL in Xenopus egg extracts to study the mechanism of lesion bypass. Deep sequencing of ICL repair products showed that the approach and extension steps are largely error-free. However, a short mutagenic tract is introduced in the vicinity of the lesion, with a maximum mutation frequency of ~1%. Our data further suggest that approach is performed by a replicative polymerase, while extension involves a complex of Rev1 and DNA polymerase ζ. Rev1-pol ζ recruitment requires the Fanconi anemia core complex but not FancI-FancD2. Our results begin to illuminate how lesion bypass is integrated with chromosomal DNA replication to limit ICL repair-associated mutagenesis.
Keywords: Fanconi anemia; genome stability; interstrand cross‐link repair; translesion synthesis.
© 2015 The Authors.
Figures

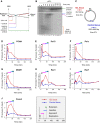
A Schematic representation of lesion bypass intermediates generated by the rightward fork after AflIII digestion of pICL.
B pICL replication intermediates were digested with AflIII and separated on a denaturing gel. Nascent strands generated by the rightward fork and extension product are shown. Analogous products are generated by the leftward fork (not shown).
C Location of primer pairs used for ChIP.
D–J Recruitment of replication and repair proteins to the ICL and the control loci. pICL was replicated, and samples were withdrawn at the indicated times for ChIP with antibodies against (D) PCNA, (E) pol δ, (F) pol ε, (G) MCM7, (H) Rev1, (I) Rev7, or (J) FancA. Experimental replicates are shown in Supplementary Fig S1. In each graph, colored bars indicate the approximate timing of TLS events (see inset for legend).
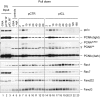

A Rev1-depleted NPE and a dilution series of mock-depleted NPE were analyzed by Western blotting using Rev1 and Rev7 antibodies. 100% corresponds to 0.25 μl of NPE.
B Reciprocal co-immunoprecipitation of Rev1 and Rev7. 20% of input (0.8 μl of NPE) and proteins precipitated from 4 μl of NPE (IP) were blotted with Rev1 and Rev7. preIm, preimmune serum.
C, D pICL was replicated in mock- or Rev1-depleted egg extracts. ChIP was performed with antibodies against Rev1 (C) and Rev7 (D) at the indicated time points. An experimental replicate of (D) is shown in Supplementary Fig S4A.
E Left: cartoon of pICL indicating the SapI site that is blocked by the ICL. Right: DNA samples from pICL replication (performed in parallel with those shown in C and D) were cut with SapI, and repair efficiency was calculated as described (Knipscheer et al, 2012). The background level of SapI-cleavable products in the Rev1-depleted reaction is due to contamination of pICL by un-cross-linked plasmid (Knipscheer et al, 2009). The amount of SapI-cleavable products can vary between experiments due to differences in the amount of un-cross-linked plasmid and in the repair capacity of individual batches of extract.
F Lesion bypass in mock- and Rev1-depleted extracts. DNA samples from (E) were cut with AflIII and analyzed on a denaturing gel, as described in Fig2B. A repetition of this experiment is shown in Supplementary Fig S4B.
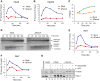
A, B pICL was replicated in mock- or FancA-depleted egg extracts, and samples were analyzed by ChIP using antibodies against FancA (A) and FancD2 (B).
C FancA depletion inhibits ICL repair. ICL repair efficiency in the extracts used in (A) and (B) was determined as in Fig4E.
D FancA depletion inhibits insertion. DNA samples from the reactions in (C) were cut with AflIII and resolved on a sequencing gel, as described in Fig2B.
E, F Rev1 and Rev7 binding to the ICL locus is reduced in FancA-depleted extract. Samples from the same reactions used in (A) and (B) were analyzed by ChIP with antibodies against Rev1 (E) and Rev7 (F). An experimental replicate is shown in Supplementary Fig S5D and E.
G Reciprocal co-immunoprecipitation of FancA and Rev1. pICL (4.2 ng/μl) was replicated in egg extract. After 40 min, 5-μl aliquots of the reaction were immunoprecipitated with the indicated antibodies. Benzonase nuclease or buffer was added during the final 15 min of the IP. 50% of input (0.3 μl of the replication reaction), supernatant (S), and precipitated proteins (P) were blotted for FancA and Rev1. preIm: preimmune serum.
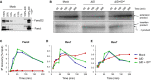
A Immunodepletion of FancD2 and FancI. A dilution series of mock-depleted NPE, FancI/FancD2-depleted NPE, and FancI/FancD2-depleted NPE supplemented with recombinant wild-type xlFancD2/FancI complex were analyzed by Western blotting using FancD2 and FancI antibodies.
B Lesion bypass in mock-depleted and FancI/FancD2-depleted extracts and in FancI–FancD2-depleted extracts supplemented with wild-type recombinant FancI–FancD2 complex was analyzed as described in Fig2B. Accumulation of the −1 product shows that the ID-depleted egg extract was functionally depleted.
C–E Extracts depleted as in (B) were analyzed by ChIP using antibodies against FancI (C), Rev1 (D), and Rev7 (E). An experimental replicate of (E) is shown in Supplementary Fig S6B.

A schematic representation of fork convergence, incisions and lesion bypass using either of the parental strands as the template. For the nascent strand (blue arrow), the nucleotide positions relative to the position of the adduct in each parental strand are indicated (red numbers).
pCTR and pICL replication products were recovered after 60 and 240 min, respectively. A 115-nt-long fragment surrounding the cross-link (present in pICL only) was deep-sequenced. A total of 1.7 million reads for pCTR and 2.6 million reads for pICL were obtained. For both plasmids, the misincorporation frequency in a 20-bp region surrounding the ICL is displayed. Nucleotide positions for the leftward and rightward forks are indicated, as in (A).
Distribution of nucleotide misincorporations in (A). The height of each colored segment represents the frequency with which that nucleotide is incorporated minus the baseline frequency in pCTR. The total height of the bars represents the overall misincorporation frequency minus baseline. Nucleotide positions for the leftward and rightward forks are indicated, as in (A).
References
-
- Albertella MR, Green CM, Lehmann AR, O’Connor MJ. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65:9799–9806. - PubMed
-
- Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–777. - PubMed
-
- Bassett E, King NM, Bryant MF, Hector S, Pendyala L, Chaney SG, Cordeiro-Stone M. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res. 2004;64:6469–6475. - PubMed
-
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

