FERAL: network-based classifier with application to breast cancer outcome prediction
- PMID: 26072498
- PMCID: PMC4765883
- DOI: 10.1093/bioinformatics/btv255
FERAL: network-based classifier with application to breast cancer outcome prediction
Abstract
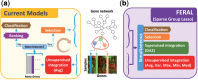
Motivation: Breast cancer outcome prediction based on gene expression profiles is an important strategy for personalize patient care. To improve performance and consistency of discovered markers of the initial molecular classifiers, network-based outcome prediction methods (NOPs) have been proposed. In spite of the initial claims, recent studies revealed that neither performance nor consistency can be improved using these methods. NOPs typically rely on the construction of meta-genes by averaging the expression of several genes connected in a network that encodes protein interactions or pathway information. In this article, we expose several fundamental issues in NOPs that impede on the prediction power, consistency of discovered markers and obscures biological interpretation.
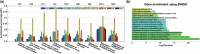
Results: To overcome these issues, we propose FERAL, a network-based classifier that hinges upon the Sparse Group Lasso which performs simultaneous selection of marker genes and training of the prediction model. An important feature of FERAL, and a significant departure from existing NOPs, is that it uses multiple operators to summarize genes into meta-genes. This gives the classifier the opportunity to select the most relevant meta-gene for each gene set. Extensive evaluation revealed that the discovered markers are markedly more stable across independent datasets. Moreover, interpretation of the marker genes detected by FERAL reveals valuable mechanistic insight into the etiology of breast cancer.
Availability and implementation: All code is available for download at: http://homepage.tudelft.nl/53a60/resources/FERAL/FERAL.zip.
© The Author 2015. Published by Oxford University Press.
Figures
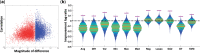




References
-
- Albert R. (2005) Scale-free networks in cell biology. J. Cell Sci. , 118, 4947–4957. - PubMed
-
- Babaei S., et al. (2011) Integrating protein family sequence similarities with gene expression to find signature gene networks in breast cancer metastasis. In: Loog M., et al. (eds), 6th IAPR International Conference, Pattern Recognition in Bioinformatics (PRIB). Springer-Verlag Berlin Heidelberg, Delft, The Netherlands, pp. 247–259.
-
- Chen G., et al. (2002) Evaluation and comparison of clustering algorithms in analyzing ES cell gene expression data. Stat. Sin. , 12, 241–262.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

