Functional advantages of dynamic protein disorder
- PMID: 26073260
- PMCID: PMC4586362
- DOI: 10.1016/j.febslet.2015.06.003
Functional advantages of dynamic protein disorder
Abstract
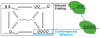
Intrinsically disordered proteins participate in many important cellular regulatory processes. The absence of a well-defined structure in the free state of a disordered domain, and even on occasion when it is bound to physiological partners, is fundamental to its function. Disordered domains are frequently the location of multiple sites for post-translational modification, the key element of metabolic control in the cell. When a disordered domain folds upon binding to a partner, the resulting complex buries a far greater surface area than in an interaction of comparably-sized folded proteins, thus maximizing specificity at modest protein size. Disorder also maintains accessibility of sites for post-translational modification. Because of their inherent plasticity, disordered domains frequently adopt entirely different structures when bound to different partners, increasing the repertoire of available interactions without the necessity for expression of many different proteins. This feature also adds to the faithfulness of cellular regulation, as the availability of a given disordered domain depends on competition between various partners relevant to different cellular processes.
Keywords: Coupled folding and binding; Post-translational modification; Transcriptional activation.
Copyright © 2015 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
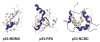


References
-
- Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–91. - PubMed
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–82. - PubMed
-
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

