Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes
- PMID: 26074426
- PMCID: PMC4490391
- DOI: 10.1038/ncomms8438
Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes
Abstract
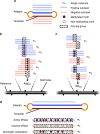
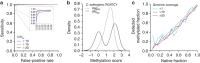
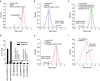
Beyond its role in host defense, bacterial DNA methylation also plays important roles in the regulation of gene expression, virulence and antibiotic resistance. Bacterial cells in a clonal population can generate epigenetic heterogeneity to increase population-level phenotypic plasticity. Single molecule, real-time (SMRT) sequencing enables the detection of N6-methyladenine and N4-methylcytosine, two major types of DNA modifications comprising the bacterial methylome. However, existing SMRT sequencing-based methods for studying bacterial methylomes rely on a population-level consensus that lacks the single-cell resolution required to observe epigenetic heterogeneity. Here, we present SMALR (single-molecule modification analysis of long reads), a novel framework for single molecule-level detection and phasing of DNA methylation. Using seven bacterial strains, we show that SMALR yields significantly improved resolution and reveals distinct types of epigenetic heterogeneity. SMALR is a powerful new tool that enables de novo detection of epigenetic heterogeneity and empowers investigation of its functions in bacterial populations.
Conflict of interest statement
E.E.S. is on the scientific advisory board of Pacific Biosciences.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA281410
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

