A Systematic Approach to Identify Markers of Distinctly Activated Human Macrophages
- PMID: 26074920
- PMCID: PMC4445387
- DOI: 10.3389/fimmu.2015.00253
A Systematic Approach to Identify Markers of Distinctly Activated Human Macrophages
Abstract
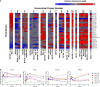
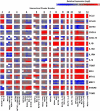
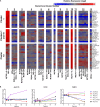
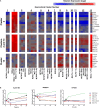
Polarization has been a useful concept for describing activated macrophage phenotypes and gene expression profiles. However, macrophage activation status within tumors and other settings are often inferred based on only a few markers. Complicating matters for relevance to human biology, many macrophage activation markers have been best characterized in mice and sometimes are not similarly regulated in human macrophages. To identify novel markers of activated human macrophages, gene expression profiles for human macrophages of a single donor subjected to 33 distinct activating conditions were obtained and a set of putative activation markers were subsequently evaluated in macrophages from multiple donors using integrated fluidic circuit (IFC)-based RT-PCR. Using unsupervised hierarchical clustering of the microarray screen, highly altered transcripts (>4-fold change in expression) sorted the macrophage transcription profiles into two major and 13 minor clusters. Among the 1874 highly altered transcripts, over 100 were uniquely altered in one major or two related minor clusters. IFC PCR-derived data confirmed the microarray results and determined the kinetics of expression of potential macrophage activation markers. Transcripts encoding chemokines, cytokines, and cell surface were prominent in our analyses. The activation markers identified by this study could be used to better characterize tumor-associated macrophages from biopsies as well as other macrophage populations collected from human clinical samples.
Keywords: activation markers; human macrophages; integrated fluidic circuit RT-PCR; macrophage polarization; microarray.
Figures

Similar articles
-
Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin.Cell Immunol. 2013 Jan;281(1):51-61. doi: 10.1016/j.cellimm.2013.01.010. Epub 2013 Feb 4. Cell Immunol. 2013. PMID: 23454681
-
Identification of interleukin-1 and interleukin-6-responsive genes in human monocyte-derived macrophages using microarrays.Biochim Biophys Acta. 2008 Jun-Jul;1779(6-7):383-9. doi: 10.1016/j.bbagrm.2008.04.006. Epub 2008 May 2. Biochim Biophys Acta. 2008. PMID: 18498781
-
Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways.Cell Signal. 2014 Feb;26(2):192-7. doi: 10.1016/j.cellsig.2013.11.004. Epub 2013 Nov 9. Cell Signal. 2014. PMID: 24219909 Review.
-
Transcriptional control of macrophage polarization.Arterioscler Thromb Vasc Biol. 2013 Jun;33(6):1135-44. doi: 10.1161/ATVBAHA.113.301453. Epub 2013 May 2. Arterioscler Thromb Vasc Biol. 2013. PMID: 23640482 Review.
-
Identification of Differentially Expressed Long Non-coding RNAs in Polarized Macrophages.Sci Rep. 2016 Jan 22;6:19705. doi: 10.1038/srep19705. Sci Rep. 2016. PMID: 26796525 Free PMC article.
Cited by
-
Macrophage nuclear receptors: Emerging key players in infectious diseases.PLoS Pathog. 2019 Mar 21;15(3):e1007585. doi: 10.1371/journal.ppat.1007585. eCollection 2019 Mar. PLoS Pathog. 2019. PMID: 30897154 Free PMC article. Review.
-
Species dependent impact of helminth-derived antigens on human macrophages infected with Mycobacterium tuberculosis: Direct effect on the innate anti-mycobacterial response.PLoS Negl Trop Dis. 2017 Feb 13;11(2):e0005390. doi: 10.1371/journal.pntd.0005390. eCollection 2017 Feb. PLoS Negl Trop Dis. 2017. PMID: 28192437 Free PMC article.
-
Metabolic Programming of Macrophages: Implications in the Pathogenesis of Granulomatous Disease.Front Immunol. 2019 Oct 4;10:2265. doi: 10.3389/fimmu.2019.02265. eCollection 2019. Front Immunol. 2019. PMID: 31681260 Free PMC article. Review.
-
Short-course radiotherapy promotes pro-inflammatory macrophages via extracellular vesicles in human rectal cancer.J Immunother Cancer. 2020 Aug;8(2):e000667. doi: 10.1136/jitc-2020-000667. J Immunother Cancer. 2020. PMID: 32817359 Free PMC article.
-
Monocyte polarization in children with falciparum malaria: relationship to nitric oxide insufficiency and disease severity.Sci Rep. 2016 Jul 7;6:29151. doi: 10.1038/srep29151. Sci Rep. 2016. PMID: 27385484 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

