Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection
- PMID: 26075598
- PMCID: PMC4468249
- DOI: 10.1371/journal.pone.0129682
Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection
Abstract
Background: Over 2.5 billion people are exposed to the risk of contracting dengue fever (DF). Early diagnosis of DF helps to diminish its burden on public health. Real-time reverse transcription polymerase amplification assays (RT-PCR) are the standard method for molecular detection of the dengue virus (DENV). Real-time RT-PCR analysis is not suitable for on-site screening since mobile devices are large, expensive, and complex. In this study, two RT-recombinase polymerase amplification (RT-RPA) assays were developed to detect DENV1-4.
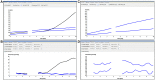
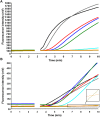
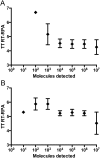
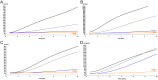
Methodology/principal findings: Using two quantitative RNA molecular standards, the analytical sensitivity of a RT-RPA targeting the 3´non-translated region of DENV1-4 was found to range from 14 (DENV4) to 241 (DENV1-3) RNA molecules detected. The assay was specific and did not cross detect other Flaviviruses. The RT-RPA assay was tested in a mobile laboratory combining magnetic-bead based total nucleic acid extraction and a portable detection device in Kedougou (Senegal) and in Bangkok (Thailand). In Kedougou, the RT-RPA was operated at an ambient temperature of 38 °C with auxiliary electricity tapped from a motor vehicle and yielded a clinical sensitivity and specificity of 98% (n=31) and 100% (n=23), respectively. While in the field trial in Bangkok, the clinical sensitivity and specificity were 72% (n=90) and 100%(n=41), respectively.
Conclusions/significance: During the first 5 days of infection, the developed DENV1-4 RT-RPA assays constitute a suitable accurate and rapid assay for DENV diagnosis. Moreover, the use of a portable fluorescence-reading device broadens its application potential to the point-of-care for outbreak investigations.
Conflict of interest statement
Figures

References
-
- WHO. Dengue fact sheets. Available: http://wwwwprowhoint/mediacentre/factsheets/fs_09032012_Dengue/en/. 2014.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

