Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- PMID: 26076345
- PMCID: PMC4468173
- DOI: 10.1371/journal.ppat.1004954
Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
Abstract
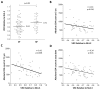
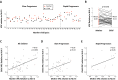
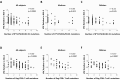
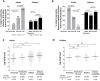
HLA class I polymorphism has a major influence on adult HIV disease progression. An important mechanism mediating this effect is the impact on viral replicative capacity (VRC) of the escape mutations selected in response to HLA-restricted CD8+ T-cell responses. Factors that contribute to slow progression in pediatric HIV infection are less well understood. We here investigate the relationship between VRC and disease progression in pediatric infection, and the effect of HLA on VRC and on disease outcome in adult and pediatric infection. Studying a South African cohort of >350 ART-naïve, HIV-infected children and their mothers, we first observed that pediatric disease progression is significantly correlated with VRC. As expected, VRCs in mother-child pairs were strongly correlated (p = 0.004). The impact of the protective HLA alleles, HLA-B*57, HLA-B*58:01 and HLA-B*81:01, resulted in significantly lower VRCs in adults (p<0.0001), but not in children. Similarly, in adults, but not in children, VRCs were significantly higher in subjects expressing the disease-susceptible alleles HLA-B*18:01/45:01/58:02 (p = 0.007). Irrespective of the subject, VRCs were strongly correlated with the number of Gag CD8+ T-cell escape mutants driven by HLA-B*57/58:01/81:01 present in each virus (p = 0.0002). In contrast to the impact of VRC common to progression in adults and children, the HLA effects on disease outcome, that are substantial in adults, are small and statistically insignificant in infected children. These data further highlight the important role that VRC plays both in adult and pediatric progression, and demonstrate that HLA-independent factors, yet to be fully defined, are predominantly responsible for pediatric non-progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

