Development of a Mild Viral Expression System for Gain-Of-Function Study of Phytoplasma Effector In Planta
- PMID: 26076458
- PMCID: PMC4468105
- DOI: 10.1371/journal.pone.0130139
Development of a Mild Viral Expression System for Gain-Of-Function Study of Phytoplasma Effector In Planta
Abstract
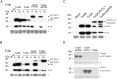
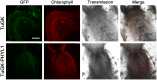
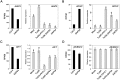
PHYL1 and SAP54 are orthologs of pathogenic effectors of Aster yellow witches'-broom (AYWB) phytoplasma and Peanut witches'-broom (PnWB) phytoplasma, respectively. These effectors cause virescence and phyllody symptoms (hereafter leafy flower) in phytoplasma-infected plants. T0 lines of transgenic Arabidopsis expressing the PHYL1 or SAP54 genes (PHYL1 or SAP54 plants) show a leafy flower phenotype and result in seedless, suggesting that PHYL1 and SAP54 interfere with reproduction stage that restrict gain-of-function studies in the next generation of transgenic plants. Turnip mosaic virus (TuMV) mild strain (TuGK) has an Arg182Lys mutation in the helper-component proteinase (HC-ProR182K) that blocks suppression of the miRNA pathway and prevents symptom development in TuGK-infected plants. We exploited TuGK as a viral vector for gain-of-function studies of PHYL1 and SAP54 in Arabidopsis plants. TuGK-PHYL1- and TuGK-SAP54-infected Arabidopsis plants produced identical leafy flower phenotypes and similar gene expression profiles as PHYL1 and SAP54 plants. In addition, the leafy flower formation rate was enhanced in TuGK-PHYL1- or TuGK-SAP54-infected Arabidopsis plants that compared with the T0 lines of PHYL1 plants. These results provide more evidence and novel directions for further studying the mechanism of PHYL1/SAP54-mediated leafy flower development. In addition, the TuGK vector is a good alternative in transgenic plant approaches for rapid gene expression in gain-of-function studies.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

