Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress
- PMID: 26077160
- PMCID: PMC4950008
- DOI: 10.1111/mmi.13091
Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress
Abstract
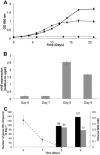
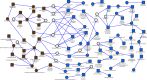
A key component to the success of Mycobacterium tuberculosis as a pathogen is the ability to sense and adapt metabolically to the diverse range of conditions encountered in vivo, such as oxygen tension, environmental pH and nutrient availability. Although nitrogen is an essential nutrient for every organism, little is known about the genes and pathways responsible for nitrogen assimilation in M. tuberculosis. In this study we have used transcriptomics and chromatin immunoprecipitation and high-throughput sequencing to address this. In response to nitrogen starvation, a total of 185 genes were significantly differentially expressed (96 up-regulated and 89 down regulated; 5% genome) highlighting several significant areas of metabolic change during nitrogen limitation such as nitrate/nitrite metabolism, aspartate metabolism and changes in cell wall biosynthesis. We identify GlnR as a regulator involved in the nitrogen response, controlling the expression of at least 33 genes in response to nitrogen limitation. We identify a consensus GlnR binding site and relate its location to known transcriptional start sites. We also show that the GlnR response regulator plays a very different role in M. tuberculosis to that in non-pathogenic mycobacteria, controlling genes involved in nitric oxide detoxification and intracellular survival instead of genes involved in nitrogen scavenging.
© 2015 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures


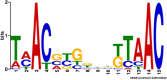
References
-
- Agarwal, N. , Lamichhane, G. , Gupta, R. , Nolan, S. , and Bishai, W.R. (2009) Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature 460: 98–102. - PubMed
-
- Amon, J. , Titgemeyer, F. , and Burkovski, A. (2009) A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J Mol Microbiol Biotechnol 17: 20–29. - PubMed
-
- Amon, J. , Titgemeyer, F. , and Burkovski, A. (2010) Common patterns – unique features: Nitrogen metabolism and regulation in Gram‐positive bacteria. FEMS Microbiol Rev 34: 588–605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

