Characterization of Three Mycobacterium spp. with Potential Use in Bioremediation by Genome Sequencing and Comparative Genomics
- PMID: 26079817
- PMCID: PMC4524478
- DOI: 10.1093/gbe/evv111
Characterization of Three Mycobacterium spp. with Potential Use in Bioremediation by Genome Sequencing and Comparative Genomics
Abstract
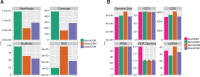
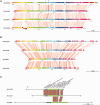
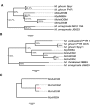
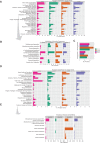
We provide the genome sequences of the type strains of the polychlorophenol-degrading Mycobacterium chlorophenolicum (DSM43826), the degrader of chlorinated aliphatics Mycobacterium chubuense (DSM44219) and Mycobacterium obuense (DSM44075) that has been tested for use in cancer immunotherapy. The genome sizes of M. chlorophenolicum, M. chubuense, and M. obuense are 6.93, 5.95, and 5.58 Mb with GC-contents of 68.4%, 69.2%, and 67.9%, respectively. Comparative genomic analysis revealed that 3,254 genes are common and we predicted approximately 250 genes acquired through horizontal gene transfer from different sources including proteobacteria. The data also showed that the biodegrading Mycobacterium spp. NBB4, also referred to as M. chubuense NBB4, is distantly related to the M. chubuense type strain and should be considered as a separate species, we suggest it to be named Mycobacterium ethylenense NBB4. Among different categories we identified genes with potential roles in: biodegradation of aromatic compounds and copper homeostasis. These are the first nonpathogenic Mycobacterium spp. found harboring genes involved in copper homeostasis. These findings would therefore provide insight into the role of this group of Mycobacterium spp. in bioremediation as well as the evolution of copper homeostasis within the Mycobacterium genus.
Keywords: Mycobacterium; biodegradation; copper homeostasis; genome sequencing; oxygenases.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Adékambi T, Drancourt M. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol. 54:2095–2105. - PubMed
-
- Agustí G, Astola O, Rodríguez-Güell E, Julián E, Luquin M. 2008. Surface spreading motility shown by a group of phylogenetically related, rapidly growing pigmented mycobacteria suggests that motility is a common property of mycobacterial species but is restricted to smooth colonies. J Bacteriol. 190:6894–6902. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

