Epidermal TRPM8 channel isoform controls the balance between keratinocyte proliferation and differentiation in a cold-dependent manner
- PMID: 26080404
- PMCID: PMC4491737
- DOI: 10.1073/pnas.1423357112
Epidermal TRPM8 channel isoform controls the balance between keratinocyte proliferation and differentiation in a cold-dependent manner
Abstract
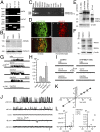
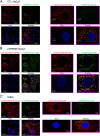
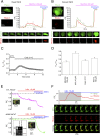
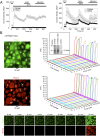
Deviation of the ambient temperature is one of the most ubiquitous stimuli that continuously affect mammals' skin. Although the role of the warmth receptors in epidermal homeostasis (EH) was elucidated in recent years, the mystery of the keratinocyte mild-cold sensor remains unsolved. Here we report the cloning and characterization of a new functional epidermal isoform of the transient receptor potential M8 (TRPM8) mild-cold receptor, dubbed epidermal TRPM8 (eTRPM8), which is localized in the keratinocyte endoplasmic reticulum membrane and controls mitochondrial Ca(2+) concentration ([Ca(2+)]m). In turn, [Ca(2+)]m modulates ATP and superoxide (O2(·-)) synthesis in a cold-dependent manner. We report that this fine tuning of ATP and O2(·-) levels by cooling controls the balance between keratinocyte proliferation and differentiation. Finally, to ascertain eTRPM8's role in EH in vivo we developed a new functional knockout mouse strain by deleting the pore domain of TRPM8 and demonstrated that eTRPM8 knockout impairs adaptation of the epidermis to low temperatures.
Keywords: calcium; cold; eTRPM8; epidermal homeostasis; mitochondria bioenergetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




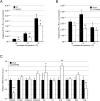

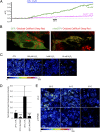
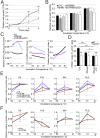


References
-
- Smith AD, Crabtree DR, Bilzon JL, Walsh NP. The validity of wireless iButtons and thermistors for human skin temperature measurement. Physiol Meas. 2010;31(1):95–114. - PubMed
-
- Lumpkin EA, Caterina MJ. Mechanisms of sensory transduction in the skin. Nature. 2007;445(7130):858–865. - PubMed
-
- Caterina MJ, Julius D. The vanilloid receptor: A molecular gateway to the pain pathway. Annu Rev Neurosci. 2001;24:487–517. - PubMed
-
- McKemy DD, Neuhausser WM, Julius D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 2002;416(6876):52–58. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

