Functional organization of the human 4D Nucleome
- PMID: 26080430
- PMCID: PMC4491792
- DOI: 10.1073/pnas.1505822112
Functional organization of the human 4D Nucleome
Abstract
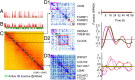
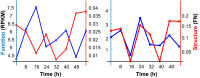
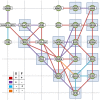
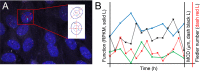
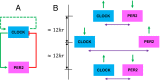
The 4D organization of the interphase nucleus, or the 4D Nucleome (4DN), reflects a dynamical interaction between 3D genome structure and function and its relationship to phenotype. We present initial analyses of the human 4DN, capturing genome-wide structure using chromosome conformation capture and 3D imaging, and function using RNA-sequencing. We introduce a quantitative index that measures underlying topological stability of a genomic region. Our results show that structural features of genomic regions correlate with function with surprising persistence over time. Furthermore, constructing genome-wide gene-level contact maps aided in identifying gene pairs with high potential for coregulation and colocalization in a manner consistent with expression via transcription factories. We additionally use 2D phase planes to visualize patterns in 4DN data. Finally, we evaluated gene pairs within a circadian gene module using 3D imaging, and found periodicity in the movement of clock circadian regulator and period circadian clock 2 relative to each other that followed a circadian rhythm and entrained with their expression.
Keywords: 4D Nucleome; Laplacian; interphase nucleus; networks; phase plane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

