Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation
- PMID: 26080448
- PMCID: PMC4491752
- DOI: 10.1073/pnas.1509658112
Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation
Abstract
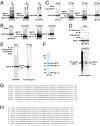
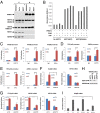
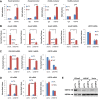
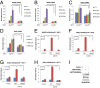
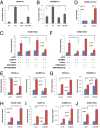
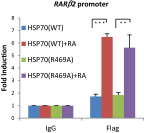
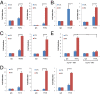
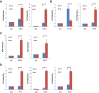
Although "histone" methyltransferases and demethylases are well established to regulate transcriptional programs and to use nonhistone proteins as substrates, their possible roles in regulation of heat-shock proteins in the nucleus have not been investigated. Here, we report that a highly conserved arginine residue, R469, in HSP70 (heat-shock protein of 70 kDa) proteins, an evolutionarily conserved protein family of ATP-dependent molecular chaperone, was monomethylated (me1), at least partially, by coactivator-associated arginine methyltransferase 1/protein arginine methyltransferase 4 (CARM1/PRMT4) and demethylated by jumonji-domain-containing 6 (JMJD6), both in vitro and in cultured cells. Functional studies revealed that HSP70 could directly regulate retinoid acid (RA)-induced retinoid acid receptor β2 (RARβ2) gene transcription through its binding to chromatin, with R469me1 being essential in this process. HSP70's function in gene transcriptional regulation appears to be distinct from its protein chaperon activity. R469me1 was shown to mediate the interaction between HSP70 and TFIIH, which involves in RNA polymerase II phosphorylation and thus transcriptional initiation. Our findings expand the repertoire of nonhistone substrates targeted by PRMT4 and JMJD6, and reveal a new function of HSP70 proteins in gene transcription at the chromatin level aside from its classic role in protein folding and quality control.
Keywords: arginine methylation; gene transcription; heat-shock proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. - PubMed
-
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148(2):558–567. - PubMed
-
- Murray K. The occurrence of epsilon-N-nethyl lysine in histones. Biochemistry. 1964;3:10–15. - PubMed
-
- Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

