Alk1 and Alk5 inhibition by Nrp1 controls vascular sprouting downstream of Notch
- PMID: 26081042
- PMCID: PMC4557308
- DOI: 10.1038/ncomms8264
Alk1 and Alk5 inhibition by Nrp1 controls vascular sprouting downstream of Notch
Abstract
Sprouting angiogenesis drives blood vessel growth in healthy and diseased tissues. Vegf and Dll4/Notch signalling cooperate in a negative feedback loop that specifies endothelial tip and stalk cells to ensure adequate vessel branching and function. Current concepts posit that endothelial cells default to the tip-cell phenotype when Notch is inactive. Here we identify instead that the stalk-cell phenotype needs to be actively repressed to allow tip-cell formation. We show this is a key endothelial function of neuropilin-1 (Nrp1), which suppresses the stalk-cell phenotype by limiting Smad2/3 activation through Alk1 and Alk5. Notch downregulates Nrp1, thus relieving the inhibition of Alk1 and Alk5, thereby driving stalk-cell behaviour. Conceptually, our work shows that the heterogeneity between neighbouring endothelial cells established by the lateral feedback loop of Dll4/Notch utilizes Nrp1 levels as the pivot, which in turn establishes differential responsiveness to TGF-β/BMP signalling.
Figures
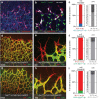
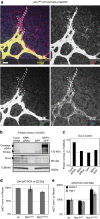
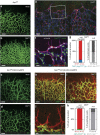
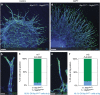
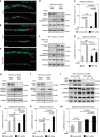

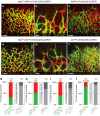
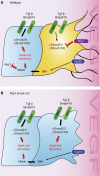
References
-
- Potente M., Gerhardt H. & Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887 (2011). - PubMed
-
- Geudens I. & Gerhardt H. Coordinating cell behaviour during blood vessel formation. Development 138, 4569–4583 (2011). - PubMed
-
- Hellstrom M. et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445, 776–780 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

