A single-residue change in the HIV-1 V3 loop associated with maraviroc resistance impairs CCR5 binding affinity while increasing replicative capacity
- PMID: 26081316
- PMCID: PMC4470041
- DOI: 10.1186/s12977-015-0177-1
A single-residue change in the HIV-1 V3 loop associated with maraviroc resistance impairs CCR5 binding affinity while increasing replicative capacity
Abstract
Background: Maraviroc (MVC) is an allosteric CCR5 inhibitor used against HIV-1 infection. While MVC-resistant viruses have been identified in patients, it still remains incompletely known how they adjust their CD4 and CCR5 binding properties to resist MVC inhibition while preserving their replicative capacity. It is thought that they maintain high efficiency of receptor binding. To date however, information about the binding affinities to receptors for inhibitor-resistant HIV-1 remains limited.
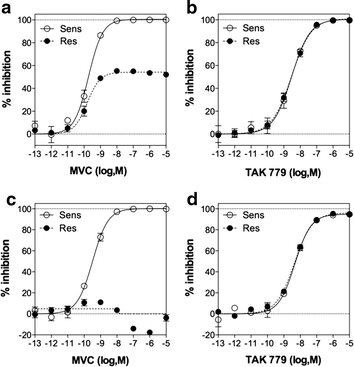
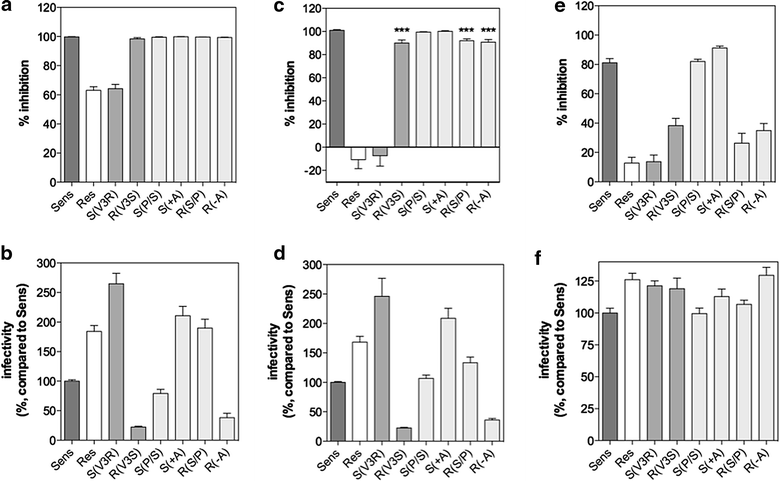
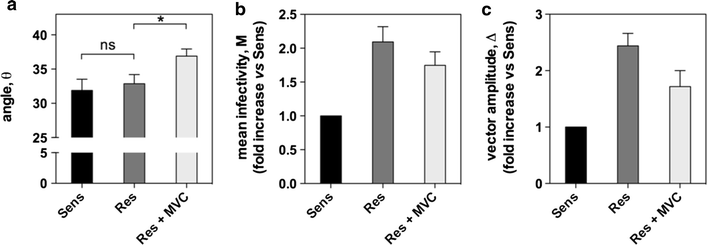
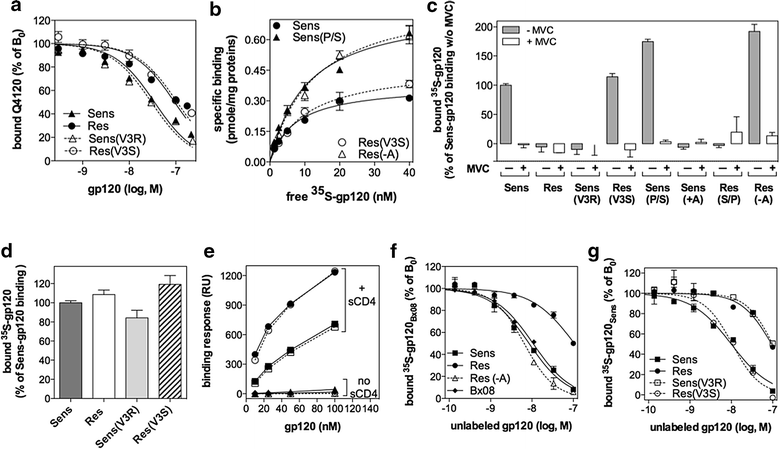
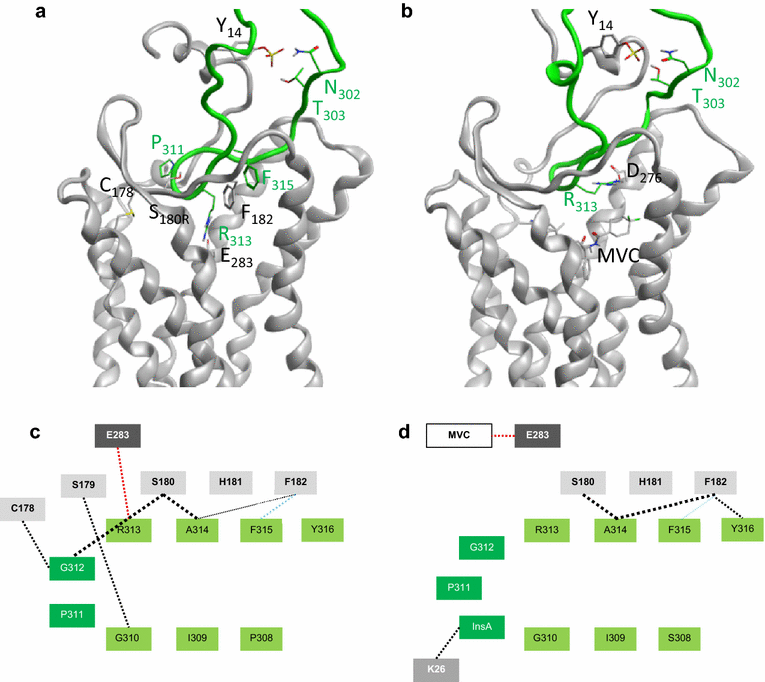
Results: Here, we show by means of viral envelope (gp120) binding experiments and virus-cell fusion kinetics that a MVC-resistant virus (MVC-Res) that had emerged as a dominant viral quasispecies in a patient displays reduced affinities for CD4 and CCR5 either free or bound to MVC, as compared to its MVC-sensitive counterpart isolated before MVC therapy. An alanine insertion within the GPG motif (G310_P311insA) of the MVC-resistant gp120 V3 loop is responsible for the decreased CCR5 binding affinity, while impaired binding to CD4 is due to sequence changes outside V3. Molecular dynamics simulations of gp120 binding to CCR5 further emphasize that the Ala insertion alters the structure of the V3 tip and weakens interaction with CCR5 ECL2. Paradoxically, infection experiments on cells expressing high levels of CCR5 also showed that Ala allows MVC-Res to use CCR5 efficiently, thereby improving viral fusion and replication efficiencies. Actually, although we found that the V3 loop of MVC-Res is required for high levels of MVC resistance, other regions outside V3 are sufficient to confer a moderate level of resistance. These sequence changes outside V3, however, come with a replication cost, which is compensated for by the Ala insertion in V3.
Conclusion: These results indicate that changes in the V3 loop of MVC-resistant viruses can augment the efficiency of CCR5-dependent steps of viral entry other than gp120 binding, thereby compensating for their decreased affinity for entry receptors and improving their fusion and replication efficiencies. This study thus sheds light on unsuspected mechanisms whereby MVC-resistant HIV-1 could emerge and grow in treated patients.
Figures



References
-
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International Union of Pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66:1–79. doi: 10.1124/pr.113.007724. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

