Whole-genome cartography of p53 response elements ranked on transactivation potential
- PMID: 26081755
- PMCID: PMC4470028
- DOI: 10.1186/s12864-015-1643-9
Whole-genome cartography of p53 response elements ranked on transactivation potential
Abstract
Background: Many recent studies using ChIP-seq approaches cross-referenced to trascriptome data and also to potentially unbiased in vitro DNA binding selection experiments are detailing with increasing precision the p53-directed gene regulatory network that, nevertheless, is still expanding. However, most experiments have been conducted in established cell lines subjected to specific p53-inducing stimuli, both factors potentially biasing the results.
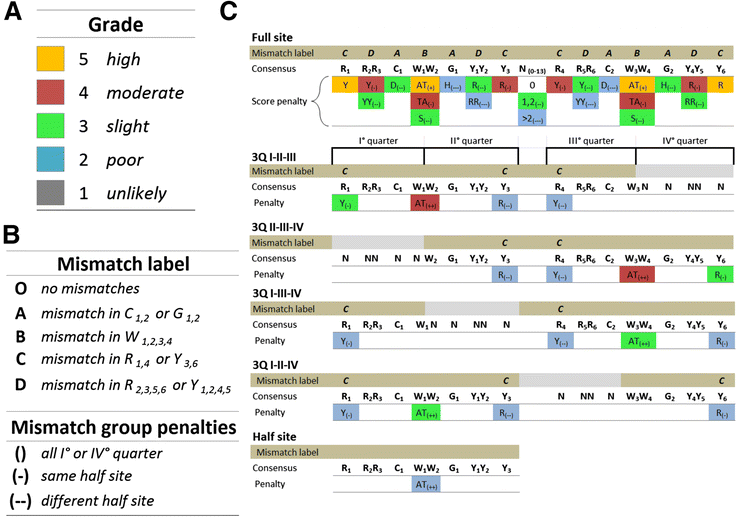
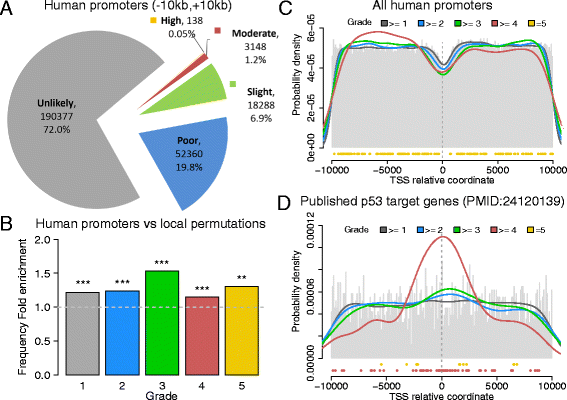
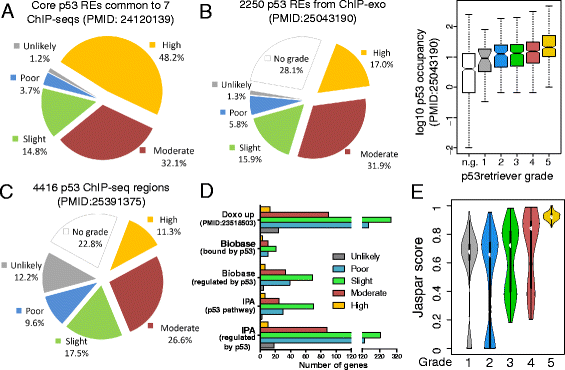
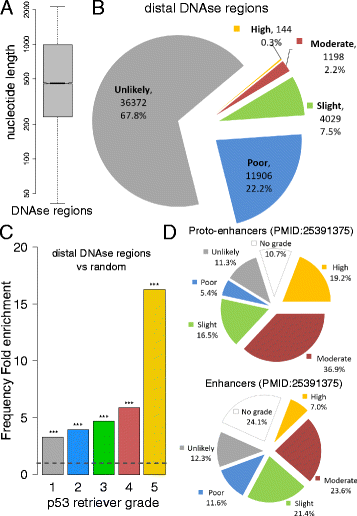
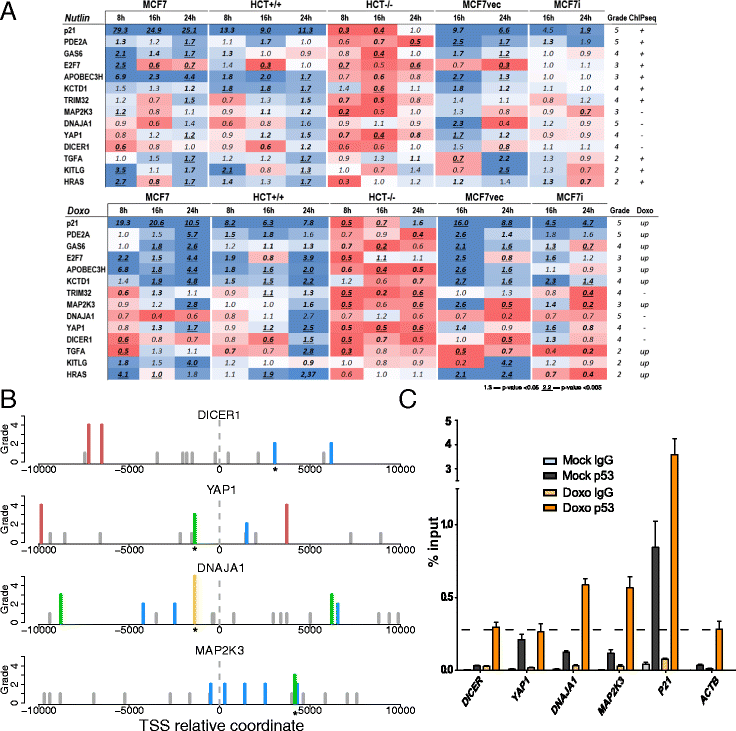
Results: We developed p53retriever, a pattern search algorithm that maps p53 response elements (REs) and ranks them according to predicted transactivation potentials in five classes. Besides canonical, full site REs, we developed specific pattern searches for non-canonical half sites and 3/4 sites and show that they can mediate p53-dependent responsiveness of associated coding sequences. Using ENCODE data, we also mapped p53 REs in about 44,000 distant enhancers and identified a 16-fold enrichment for high activity REs within those sites in the comparison with genomic regions near transcriptional start sites (TSS). Predictions from our pattern search were cross-referenced to ChIP-seq, ChIP-exo, expression, and various literature data sources. Based on the mapping of predicted functional REs near TSS, we examined expression changes of thirteen genes as a function of different p53-inducing conditions, providing further evidence for PDE2A, GAS6, E2F7, APOBEC3H, KCTD1, TRIM32, DICER, HRAS, KITLG and TGFA p53-dependent regulation, while MAP2K3, DNAJA1 and potentially YAP1 were identified as new direct p53 target genes.
Conclusions: We provide a comprehensive annotation of canonical and non-canonical p53 REs in the human genome, ranked on predicted transactivation potential. We also establish or corroborate direct p53 transcriptional control of thirteen genes. The entire list of identified and functionally classified p53 REs near all UCSC-annotated genes and within ENCODE mapped enhancer elements is provided. Our approach is distinct from, and complementary to, existing methods designed to identify p53 response elements. p53retriever is available as an R package at: http://tomateba.github.io/p53retriever .
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

