Small molecule/ML327 mediated transcriptional de-repression of E-cadherin and inhibition of epithelial-to-mesenchymal transition
- PMID: 26082441
- PMCID: PMC4673210
- DOI: 10.18632/oncotarget.4473
Small molecule/ML327 mediated transcriptional de-repression of E-cadherin and inhibition of epithelial-to-mesenchymal transition
Abstract
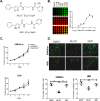
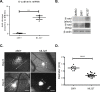
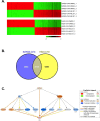
Transcriptional repression of E-cadherin is a hallmark of Epithelial-to-Mesenchymal Transition (EMT) and is associated with cancer cell invasion and metastasis. Understanding the mechanisms underlying E-cadherin repression during EMT may provide insights into the development of novel targeted therapeutics for cancer. Here, we report on the chemical probe, ML327, which de-represses E-cadherin transcription, partially reverses EMT, and inhibits cancer cell invasiveness and tumor cell migration in vitro and in vivo. Induction of E-cadherin mRNA expression by ML327 treatment does not require de novo protein synthesis. RNA sequencing analysis revealed that ML327 treatment significantly alters expression of over 2,500 genes within three hours in the presence of the translational inhibitor, cycloheximide. Network analysis reveals Hepatocyte Nuclear Factor 4-alpha (HNF4α) as the most significant upstream transcriptional regulator of multiple genes whose expressions were altered by ML327 treatment. Further, small interfering RNA-mediated depletion of HNF4α markedly attenuates the E-cadherin expression response to ML327. In summary, ML327 represents a valuable tool to understand mechanisms of EMT and may provide the basis for a novel targeted therapeutic strategy for carcinomas.
Keywords: E-cadherin; EMT; small molecule.
Conflict of interest statement
None.
Figures




References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57:43–66. - PubMed
-
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. - PubMed
-
- Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. - PubMed
-
- Schneider MR, Kolligs FT. E-cadherin's role in development, tissue homeostasis and disease: Insights from mouse models: Tissue-specific inactivation of the adhesion protein E-cadherin in mice reveals its functions in health and disease. Bioessays. 2015;37:294–304. - PubMed
-
- Guilford PJ, Hopkins JB, Grady WM, Markowitz SD, Willis J, Lynch H, Rajput A, Wiesner GL, Lindor NM, Burgart LJ, Toro TT, Lee D, Limacher JM, Shaw DW, Findlay MP, Reeve AE. E-cadherin germline mutations define an inherited cancer syndrome dominated by diffuse gastric cancer. Human mutation. 1999;14:249–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

