Modulation of the expression of mimivirus-encoded translation-related genes in response to nutrient availability during Acanthamoeba castellanii infection
- PMID: 26082761
- PMCID: PMC4450173
- DOI: 10.3389/fmicb.2015.00539
Modulation of the expression of mimivirus-encoded translation-related genes in response to nutrient availability during Acanthamoeba castellanii infection
Abstract
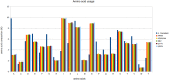
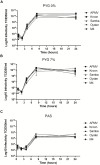
The complexity of giant virus genomes is intriguing, especially the presence of genes encoding components of the protein translation machinery such as transfer RNAs and aminoacyl-tRNA-synthetases; these features are uncommon among other viruses. Although orthologs of these genes are codified by their hosts, one can hypothesize that having these translation-related genes might represent a gain of fitness during infection. Therefore, the aim of this study was to evaluate the expression of translation-related genes by mimivirus during infection of Acanthamoeba castellanii under different nutritional conditions. In silico analysis of amino acid usage revealed remarkable differences between the mimivirus isolates and the A. castellanii host. Relative expression analysis by quantitative PCR revealed that mimivirus was able to modulate the expression of eight viral translation-related genes according to the amoebal growth condition, with a higher induction of gene expression under starvation. Some mimivirus isolates presented differences in translation-related gene expression; notably, polymorphisms in the promoter regions correlated with these differences. Two mimivirus isolates did not encode the tryptophanyl-tRNA in their genomes, which may be linked with low conservation pressure based on amino acid usage analysis. Taken together, our data suggest that mimivirus can modulate the expression of translation-related genes in response to nutrient availability in the host cell, allowing the mimivirus to adapt to different hosts growing under different nutritional conditions.
Keywords: aminoacyl-tRNA-synthetases; gene expression; mimivirus; tRNA; translation.
Figures


Similar articles
-
Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS.J Virol. 2007 Nov;81(22):12406-17. doi: 10.1128/JVI.01107-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855524 Free PMC article.
-
Experimental Analysis of Mimivirus Translation Initiation Factor 4a Reveals Its Importance in Viral Protein Translation during Infection of Acanthamoeba polyphaga.J Virol. 2018 Apr 27;92(10):e00337-18. doi: 10.1128/JVI.00337-18. Print 2018 May 15. J Virol. 2018. PMID: 29514904 Free PMC article.
-
Codon usage, amino acid usage, transfer RNA and amino-acyl-tRNA synthetases in Mimiviruses.Intervirology. 2013;56(6):364-75. doi: 10.1159/000354557. Epub 2013 Oct 17. Intervirology. 2013. PMID: 24157883
-
Mimivirus.Curr Top Microbiol Immunol. 2009;328:89-121. doi: 10.1007/978-3-540-68618-7_3. Curr Top Microbiol Immunol. 2009. PMID: 19216436 Review.
-
Mimivirus and the emerging concept of "giant" virus.Virus Res. 2006 Apr;117(1):133-44. doi: 10.1016/j.virusres.2006.01.008. Epub 2006 Feb 15. Virus Res. 2006. PMID: 16469402 Review.
Cited by
-
Assessing the biogeography of marine giant viruses in four oceanic transects.bioRxiv [Preprint]. 2023 Feb 1:2023.01.30.526306. doi: 10.1101/2023.01.30.526306. bioRxiv. 2023. Update in: ISME Commun. 2023 Apr 29;3(1):43. doi: 10.1038/s43705-023-00252-6. PMID: 36778472 Free PMC article. Updated. Preprint.
-
Giant virus biology and diversity in the era of genome-resolved metagenomics.Nat Rev Microbiol. 2022 Dec;20(12):721-736. doi: 10.1038/s41579-022-00754-5. Epub 2022 Jul 28. Nat Rev Microbiol. 2022. PMID: 35902763 Review.
-
High Transcriptional Activity and Diverse Functional Repertoires of Hundreds of Giant Viruses in a Coastal Marine System.mSystems. 2021 Aug 31;6(4):e0029321. doi: 10.1128/mSystems.00293-21. Epub 2021 Jul 13. mSystems. 2021. PMID: 34254826 Free PMC article.
-
Niemeyer Virus: A New Mimivirus Group A Isolate Harboring a Set of Duplicated Aminoacyl-tRNA Synthetase Genes.Front Microbiol. 2015 Nov 10;6:1256. doi: 10.3389/fmicb.2015.01256. eCollection 2015. Front Microbiol. 2015. PMID: 26635738 Free PMC article.
-
Assessing the biogeography of marine giant viruses in four oceanic transects.ISME Commun. 2023 Apr 29;3(1):43. doi: 10.1038/s43705-023-00252-6. ISME Commun. 2023. PMID: 37120676 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

