The orisome: structure and function
- PMID: 26082765
- PMCID: PMC4451416
- DOI: 10.3389/fmicb.2015.00545
The orisome: structure and function
Abstract
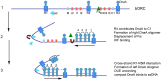
During the cell division cycle of all bacteria, DNA-protein complexes termed orisomes trigger the onset of chromosome duplication. Orisome assembly is both staged and stringently regulated to ensure that DNA synthesis begins at a precise time and only once at each origin per cycle. Orisomes comprise multiple copies of the initiator protein DnaA, which oligomerizes after interacting with specifically positioned recognition sites in the unique chromosomal replication origin, oriC. Since DnaA is highly conserved, it is logical to expect that all bacterial orisomes will share fundamental attributes. Indeed, although mechanistic details remain to be determined, all bacterial orisomes are capable of unwinding oriC DNA and assisting with loading of DNA helicase onto the single-strands. However, comparative analysis of oriCs reveals that the arrangement and number of DnaA recognition sites is surprisingly variable among bacterial types, suggesting there are many paths to produce functional orisome complexes. Fundamental questions exist about why these different paths exist and which features of orisomes must be shared among diverse bacterial types. In this review we present the current understanding of orisome assembly and function in Escherichia coli and compare the replication origins among the related members of the Gammaproteobacteria. From this information we propose that the diversity in orisome assembly reflects both the requirement to regulate the conformation of origin DNA as well as to provide an appropriate cell cycle timing mechanism that reflects the lifestyle of the bacteria. We suggest that identification of shared steps in orisome assembly may reveal particularly good targets for new antibiotics.
Keywords: DNA binding proteins; DNA replication; DnaA; oriC; orisomes; pre-replication complexes; replication origin.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

