Purification and proteomics of pathogen-modified vacuoles and membranes
- PMID: 26082896
- PMCID: PMC4451638
- DOI: 10.3389/fcimb.2015.00048
Purification and proteomics of pathogen-modified vacuoles and membranes
Abstract
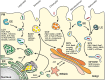
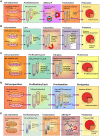
Certain pathogenic bacteria adopt an intracellular lifestyle and proliferate in eukaryotic host cells. The intracellular niche protects the bacteria from cellular and humoral components of the mammalian immune system, and at the same time, allows the bacteria to gain access to otherwise restricted nutrient sources. Yet, intracellular protection and access to nutrients comes with a price, i.e., the bacteria need to overcome cell-autonomous defense mechanisms, such as the bactericidal endocytic pathway. While a few bacteria rupture the early phagosome and escape into the host cytoplasm, most intracellular pathogens form a distinct, degradation-resistant and replication-permissive membranous compartment. Intracellular bacteria that form unique pathogen vacuoles include Legionella, Mycobacterium, Chlamydia, Simkania, and Salmonella species. In order to understand the formation of these pathogen niches on a global scale and in a comprehensive and quantitative manner, an inventory of compartment-associated host factors is required. To this end, the intact pathogen compartments need to be isolated, purified and biochemically characterized. Here, we review recent progress on the isolation and purification of pathogen-modified vacuoles and membranes, as well as their proteomic characterization by mass spectrometry and different validation approaches. These studies provide the basis for further investigations on the specific mechanisms of pathogen-driven compartment formation.
Keywords: Chlamydia; Legionella; Mycobacterium; Salmonella; Simkania; host-pathogen interactions; immuno-magnetic purification; pathogen vacuole.
Figures

References
-
- Ansong C., Wu S., Meng D., Liu X., Brewer H. M., Deatherage Kaiser B. L., et al. . (2013). Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 10153–10158. 10.1073/pnas.1221210110 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

