MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors
- PMID: 26083154
- PMCID: PMC4615225
- DOI: 10.1080/15476286.2015.1050577
MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors
Abstract
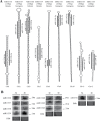
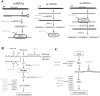
MicroRNAs (miRNAs) are small non-coding RNAs that have important regulatory functions in plant growth, development, and response to abiotic stress. Increasing evidence also supports that plant miRNAs contribute to immune responses to pathogens. Here, we used deep sequencing of small RNA libraries for global identification of rice miRNAs that are regulated by fungal elicitors. We also describe 9 previously uncharacterized miRNAs in rice. Combined small RNA and degradome analyses revealed regulatory networks enriched in elicitor-regulated miRNAs supported by the identification of their corresponding target genes. Specifically, we identified an important number of miRNA/target gene pairs involved in small RNA pathways, including miRNA, heterochromatic and trans-acting siRNA pathways. We present evidence for miRNA/target gene pairs implicated in hormone signaling and cross-talk among hormone pathways having great potential in regulating rice immunity. Furthermore, we describe miRNA-mediated regulation of Conserved-Peptide upstream Open Reading Frame (CPuORF)-containing genes in rice, which suggests the existence of a novel regulatory network that integrates miRNA and CPuORF functions in plants. The knowledge gained in this study will help in understanding the underlying regulatory mechanisms of miRNAs in rice immunity and develop appropriate strategies for rice protection.
Keywords: Magnaporthe oryzae; conserved peptide upstream open reading frame; degradome; elicitors; miRNAs; rice.
Figures







References
-
- Baulcombe D. RNA silencing in plants. Nature 2004; 431:356-63; PMID:15372043; http://dx.doi.org/10.1038/nature02874 - DOI - PubMed
-
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 2006; 20:759-71; PMID:16600909; http://dx.doi.org/10.1101/gad.1410506 - DOI - PubMed
-
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science 2003; 301:336-8; PMID:12869753; http://dx.doi.org/10.1126/science.1085242 - DOI - PubMed
-
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009; 136:669-87; PMID:19239888; http://dx.doi.org/10.1016/j.cell.2009.01.046 - DOI - PubMed
-
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 2013; 64:137-59; PMID:23330790; http://dx.doi.org/10.1146/annurev-arplant-050312-120043 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
