miRNA-26b Overexpression in Ulcerative Colitis-associated Carcinogenesis
- PMID: 26083618
- PMCID: PMC4603667
- DOI: 10.1097/MIB.0000000000000453
miRNA-26b Overexpression in Ulcerative Colitis-associated Carcinogenesis
Abstract
Background: Longstanding ulcerative colitis (UC) bears a high risk for development of UC-associated colorectal carcinoma (UCC). The inflammatory microenvironment influences microRNA expression, which in turn deregulates target gene expression. microRNA-26b (miR-26b) was shown to be instrumental in normal tissue growth and differentiation. Thus, we aimed to investigate the impact of miR-26b in inflammation-associated colorectal carcinogenesis.
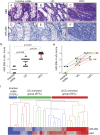
Methods: Two different cohorts of patients were investigated. In the retrospective group, a tissue microarray with 38 samples from 17 UC/UCC patients was used for miR-26b in situ hybridization and quantitative reverse transcription polymerase chain reaction analyses. In the prospective group, we investigated miR-26b expression in 25 fresh-frozen colon biopsies and corresponding serum samples of 6 UC and 15 non-UC patients, respectively. In silico analysis, Ago2-RNA immunoprecipitation, luciferase reporter assay, quantitative reverse transcription polymerase chain reaction examination, and miR-26b mimic overexpression were employed for target validation.
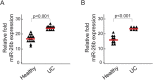
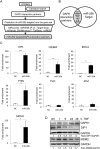
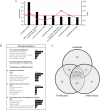
Results: miR-26b expression was shown to be upregulated with disease progression in tissues and serum of UC and UCC patients. Using miR-26b and Ki-67 expression levels, an UCC was predicted with high accuracy. We identified 4 novel miR-26b targets (DIP1, MDM2, CREBBP, BRCA1). Among them, the downregulation of the E3 ubiquitin ligase DIP1 was closely related to death-associated protein kinase stabilization along the normal mucosa-UC-UCC sequence. In silico functional pathway analysis revealed that the common cellular pathways affected by miR-26b are highly related to cancerogenesis and the development of gastrointestinal diseases.
Conclusions: We suggest that miR-26b could serve as a biomarker for inflammation-associated processes in the gastrointestinal system. Because miR-26b expression is downregulated in sporadic colon cancer, it could discriminate between UCC and the sporadic cancer type.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


Comment in
-
Clinical Value of miR-26b Discriminating Ulcerative Colitis-associated Colorectal Cancer in the Subgroup of Patients with Metastatic Disease.Inflamm Bowel Dis. 2015 Oct;21(10):E24-5. doi: 10.1097/MIB.0000000000000572. Inflamm Bowel Dis. 2015. PMID: 26308439 No abstract available.
References
-
- Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. - PubMed
-
- Bernstein CN, Blanchard JF, Kliewer E, et al. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer. 2001;91:854–862. - PubMed
-
- Habermann J, Lenander C, Roblick UJ, et al. Ulcerative colitis and colorectal carcinoma: DNA-profile, laminin-5 gamma2 chain and cyclin A expression as early markers for risk assessment. Scand J Gastroenterol. 2001;36:751–758. - PubMed
-
- Terzic J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e2105. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

