Rapid, Sensitive, and Accurate Evaluation of Drug Resistant Mutant (NS5A-Y93H) Strain Frequency in Genotype 1b HCV by Invader Assay
- PMID: 26083687
- PMCID: PMC4470996
- DOI: 10.1371/journal.pone.0130022
Rapid, Sensitive, and Accurate Evaluation of Drug Resistant Mutant (NS5A-Y93H) Strain Frequency in Genotype 1b HCV by Invader Assay
Abstract
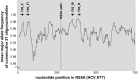
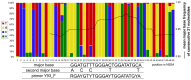
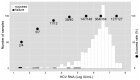
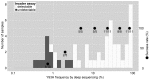
Daclatasvir and asunaprevir dual oral therapy is expected to achieve high sustained virological response (SVR) rates in patients with HCV genotype 1b infection. However, presence of the NS5A-Y93H substitution at baseline has been shown to be an independent predictor of treatment failure for this regimen. By using the Invader assay, we developed a system to rapidly and accurately detect the presence of mutant strains and evaluate the proportion of patients harboring a pre-treatment Y93H mutation. This assay system, consisting of nested PCR followed by Invader reaction with well-designed primers and probes, attained a high overall assay success rate of 98.9% among a total of 702 Japanese HCV genotype 1b patients. Even in serum samples with low HCV titers, more than half of the samples could be successfully assayed. Our assay system showed a better lower detection limit of Y93H proportion than using direct sequencing, and Y93H frequencies obtained by this method correlated well with those of deep-sequencing analysis (r = 0.85, P <0.001). The proportion of the patients with the mutant strain estimated by this assay was 23.6% (164/694). Interestingly, patients with the Y93H mutant strain showed significantly lower ALT levels (p=8.8 x 10-4), higher serum HCV RNA levels (p=4.3 x 10-7), and lower HCC risk (p=6.9 x 10-3) than those with the wild type strain. Because the method is both sensitive and rapid, the NS5A-Y93H mutant strain detection system established in this study may provide important pre-treatment information valuable not only for treatment decisions but also for prediction of disease progression in HCV genotype 1b patients.
Conflict of interest statement
Figures



References
-
- Shepard CW, Finelli L, Alter MJ Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 2005; 5: 558–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

