Molecular organization and comparative analysis of chromosome 5B of the wild wheat ancestor Triticum dicoccoides
- PMID: 26084265
- PMCID: PMC4471722
- DOI: 10.1038/srep10763
Molecular organization and comparative analysis of chromosome 5B of the wild wheat ancestor Triticum dicoccoides
Abstract
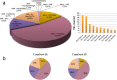
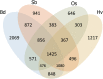
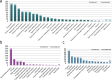
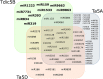
Wild emmer wheat, Triticum turgidum ssp. dicoccoides is the wild relative of Triticum turgidum, the progenitor of durum and bread wheat, and maintains a rich allelic diversity among its wild populations. The lack of adequate genetic and genomic resources, however, restricts its exploitation in wheat improvement. Here, we report next-generation sequencing of the flow-sorted chromosome 5B of T. dicoccoides to shed light into its genome structure, function and organization by exploring the repetitive elements, protein-encoding genes and putative microRNA and tRNA coding sequences. Comparative analyses with its counterparts in modern and wild wheats suggest clues into the B-genome evolution. Syntenic relationships of chromosome 5B with the model grasses can facilitate further efforts for fine-mapping of traits of interest. Mapping of 5B sequences onto the root transcriptomes of two additional T. dicoccoides genotypes, with contrasting drought tolerances, revealed several thousands of single nucleotide polymorphisms, of which 584 shared polymorphisms on 228 transcripts were specific to the drought-tolerant genotype. To our knowledge, this study presents the largest genomics resource currently available for T. dicoccoides, which, we believe, will encourage the exploitation of its genetic and genomic potential for wheat improvement to meet the increasing demand to feed the world.
Figures

References
-
- Feuillet C., Langridge P. & Waugh R. Cereal breeding takes a walk on the wild side. Trends Genet. 24, 24–32 (2008). - PubMed
-
- Nevo E. & Chen G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant. Cell Environ. 33, 670–85 (2010). - PubMed
-
- Paux E., Sourdille P., Mackay I. & Feuillet C. Sequence-based marker development in wheat: advances and applications to breeding. Biotechnol. Adv. 30, 1071–88 (2012). - PubMed
-
- Marcussen T. et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092–1250092 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

