International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors
- PMID: 26084539
- PMCID: PMC4485016
- DOI: 10.1124/pr.114.010249
International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors
Abstract
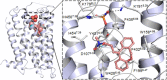
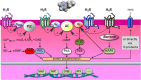
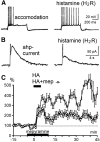
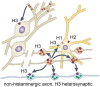
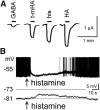
Histamine is a developmentally highly conserved autacoid found in most vertebrate tissues. Its physiological functions are mediated by four 7-transmembrane G protein-coupled receptors (H1R, H2R, H3R, H4R) that are all targets of pharmacological intervention. The receptors display molecular heterogeneity and constitutive activity. H1R antagonists are long known antiallergic and sedating drugs, whereas the H2R was identified in the 1970s and led to the development of H2R-antagonists that revolutionized stomach ulcer treatment. The crystal structure of ligand-bound H1R has rendered it possible to design new ligands with novel properties. The H3R is an autoreceptor and heteroreceptor providing negative feedback on histaminergic and inhibition on other neurons. A block of these actions promotes waking. The H4R occurs on immuncompetent cells and the development of anti-inflammatory drugs is anticipated.
Copyright © 2015 by The American Society for Pharmacology and Experimental Therapeutics.
Figures
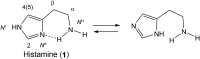
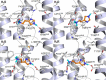





References
-
- Adami M, Pozzoli C, Menozzi A, Bertini S, Passeri B, Cantoni AM, Smits R, de Esch I, Leurs R, Coruzzi G. (2012) Effects of histamine H4 receptor ligands in a mouse model of gastric ulceration. Pharmacology 89:287–294. - PubMed
-
- Aldi S, Takano K, Tomita K, Koda K, Chan NY-K, Marino A, Salazar-Rodriguez M, Thurmond RL, Levi R. (2014) Histamine H4-receptors inhibit mast cell renin release in ischemia/reperfusion via protein kinase C ε-dependent aldehyde dehydrogenase type-2 activation. J Pharmacol Exp Ther 349:508–517. - PMC - PubMed
-
- Alewijnse AE, Smit MJ, Hoffmann M, Verzijl D, Timmerman H, Leurs R. (1998) Constitutive activity and structural instability of the wild-type human H2 receptor. J Neurochem 71:799–807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

