A unified gene catalog for the laboratory mouse reference genome
- PMID: 26084703
- PMCID: PMC4534496
- DOI: 10.1007/s00335-015-9571-1
A unified gene catalog for the laboratory mouse reference genome
Abstract
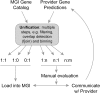
We report here a semi-automated process by which mouse genome feature predictions and curated annotations (i.e., genes, pseudogenes, functional RNAs, etc.) from Ensembl, NCBI and Vertebrate Genome Annotation database (Vega) are reconciled with the genome features in the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org) into a comprehensive and non-redundant catalog. Our gene unification method employs an algorithm (fjoin--feature join) for efficient detection of genome coordinate overlaps among features represented in two annotation data sets. Following the analysis with fjoin, genome features are binned into six possible categories (1:1, 1:0, 0:1, 1:n, n:1, n:m) based on coordinate overlaps. These categories are subsequently prioritized for assessment of annotation equivalencies and differences. The version of the unified catalog reported here contains more than 59,000 entries, including 22,599 protein-coding coding genes, 12,455 pseudogenes, and 24,007 other feature types (e.g., microRNAs, lincRNAs, etc.). More than 23,000 of the entries in the MGI gene catalog have equivalent gene models in the annotation files obtained from NCBI, Vega, and Ensembl. 12,719 of the features are unique to NCBI relative to Ensembl/Vega; 11,957 are unique to Ensembl/Vega relative to NCBI, and 3095 are unique to MGI. More than 4000 genome features fall into categories that require manual inspection to resolve structural differences in the gene models from different annotation sources. Using the MGI unified gene catalog, researchers can easily generate a comprehensive report of mouse genome features from a single source and compare the details of gene and transcript structure using MGI's mouse genome browser.
Figures






References
-
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Burger A, Bult CJ, Bushell W, Collins FS, Desaintes C, Doe B, Economides A, Eppig JT, Finnell RH, Fletcher C, Fray M, Frendewey D, Friedel RH, Grosveld FG, Hansen J, Herault Y, Hicks G, Horlein A, Houghton R, Hrabe de Angelis M, Huylebroeck D, Iyer V, de Jong PJ, Kadin JA, Kaloff C, Kennedy K, Koutsourakis M, Lloyd KC, Marschall S, Mason J, McKerlie C, McLeod MP, von Melchner H, Moore M, Mujica AO, Nagy A, Nefedov M, Nutter LM, Pavlovic G, Peterson JL, Pollock J, Ramirez-Solis R, Rancourt DE, Raspa M, Remacle JE, Ringwald M, Rosen B, Rosenthal N, Rossant J, Ruiz Noppinger P, Ryder E, Schick JZ, Schnutgen F, Schofield P, Seisenberger C, Selloum M, Simpson EM, Skarnes WC, Smedley D, Stanford WL, Stewart AF, Stone K, Swan K, Tadepally H, Teboul L, Tocchini-Valentini GP, Valenzuela D, West AP, Yamamura K, Yoshinaga Y, Wurst W. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

