Uncovering major genomic features of essential genes in Bacteria and a methanogenic Archaea
- PMID: 26084810
- PMCID: PMC4558319
- DOI: 10.1111/febs.13350
Uncovering major genomic features of essential genes in Bacteria and a methanogenic Archaea
Abstract
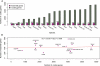
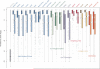
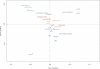
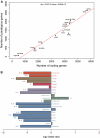
Identification of essential genes is critical to understanding the physiology of a species, proposing novel drug targets and uncovering minimal gene sets required for life. Although essential gene sets of several organisms have been determined using large-scale mutagenesis techniques, systematic studies addressing their conservation, genomic context and functions remain scant. Here we integrate 17 essential gene sets from genome-wide in vitro screenings and three gene collections required for growth in vivo, encompassing 15 Bacteria and one Archaea. We refine and generalize important theories proposed using Escherichia coli. Essential genes are typically monogenic and more conserved than nonessential genes. Genes required in vivo are less conserved than those essential in vitro, suggesting that more divergent strategies are deployed when the organism is stressed by the host immune system and unstable nutrient availability. We identified essential analogous pathways that would probably be missed by orthology-based essentiality prediction strategies. For example, Streptococcus sanguinis carries horizontally transferred isoprenoid biosynthesis genes that are widespread in Archaea. Genes specifically essential in Mycobacterium tuberculosis and Burkholderia pseudomallei are reported as potential drug targets. Moreover, essential genes are not only preferentially located in operons, but also occupy the first position therein, supporting the influence of their regulatory regions in driving transcription of whole operons. Finally, these important genomic features are shared between Bacteria and at least one Archaea, suggesting that high order properties of gene essentiality and genome architecture were probably present in the last universal common ancestor or evolved independently in the prokaryotic domains.
Keywords: essential genes; genome evolution; genome organization; operons; prokaryotes; transposon mutagenesis.
© 2015 FEBS.
Figures

Similar articles
-
High-throughput transposon mutagenesis in the family Enterobacteriaceae reveals core essential genes and rapid turnover of essentiality.mBio. 2024 Oct 16;15(10):e0179824. doi: 10.1128/mbio.01798-24. Epub 2024 Aug 29. mBio. 2024. PMID: 39207104 Free PMC article.
-
The repertoire of DNA-binding transcription factors in prokaryotes: functional and evolutionary lessons.Sci Prog. 2012;95(Pt 3):315-29. doi: 10.3184/003685012X13420097673409. Sci Prog. 2012. PMID: 23094327 Free PMC article. Review.
-
Long range segmentation of prokaryotic genomes by gene age and functionality.Nucleic Acids Res. 2024 Oct 14;52(18):11045-11059. doi: 10.1093/nar/gkae745. Nucleic Acids Res. 2024. PMID: 39193895 Free PMC article.
-
Genome-Wide Essentiality Analysis of Mycobacterium abscessus by Saturated Transposon Mutagenesis and Deep Sequencing.mBio. 2021 Jun 29;12(3):e0104921. doi: 10.1128/mBio.01049-21. Epub 2021 Jun 15. mBio. 2021. PMID: 34126767 Free PMC article.
-
Ancient origin of the tryptophan operon and the dynamics of evolutionary change.Microbiol Mol Biol Rev. 2003 Sep;67(3):303-42, table of contents. doi: 10.1128/MMBR.67.3.303-342.2003. Microbiol Mol Biol Rev. 2003. PMID: 12966138 Free PMC article. Review.
Cited by
-
Variability of Bacterial Essential Genes Among Closely Related Bacteria: The Case of Escherichia coli.Front Microbiol. 2018 May 29;9:1059. doi: 10.3389/fmicb.2018.01059. eCollection 2018. Front Microbiol. 2018. PMID: 29910775 Free PMC article. Review.
-
Genome-wide fitness analysis identifies genes required for in vitro growth and macrophage infection by African and global epidemic pathovariants of Salmonella enterica Enteritidis.Microb Genom. 2023 May;9(5):mgen001017. doi: 10.1099/mgen.0.001017. Microb Genom. 2023. PMID: 37219927 Free PMC article.
-
A deep ensemble framework for human essential gene prediction by integrating multi-omics data.Sci Rep. 2025 Jul 21;15(1):26407. doi: 10.1038/s41598-025-99164-9. Sci Rep. 2025. PMID: 40691502 Free PMC article.
-
Immunoinformatics-Aided Design and Evaluation of a Potential Multi-Epitope Vaccine against Klebsiella Pneumoniae.Vaccines (Basel). 2019 Aug 12;7(3):88. doi: 10.3390/vaccines7030088. Vaccines (Basel). 2019. PMID: 31409021 Free PMC article.
-
Large-Scale Analysis of the Mycoplasma bovis Genome Identified Non-essential, Adhesion- and Virulence-Related Genes.Front Microbiol. 2019 Sep 13;10:2085. doi: 10.3389/fmicb.2019.02085. eCollection 2019. Front Microbiol. 2019. PMID: 31572317 Free PMC article.
References
-
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nature reviews Microbiology. 2012;10:13–26. - PubMed
-
- Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT. Microbial biogeography: putting microorganisms on the map. Nature reviews Microbiology. 2006;4:102–12. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

