A Three-protein Charge Zipper Stabilizes a Complex Modulating Bacterial Gene Silencing
- PMID: 26085102
- PMCID: PMC4571853
- DOI: 10.1074/jbc.M114.630400
A Three-protein Charge Zipper Stabilizes a Complex Modulating Bacterial Gene Silencing
Abstract
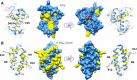
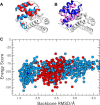
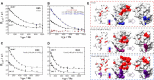
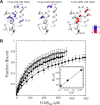
The Hha/YmoA nucleoid-associated proteins help selectively silence horizontally acquired genetic material, including pathogenicity and antibiotic resistance genes and their maintenance in the absence of selective pressure. Members of the Hha family contribute to gene silencing by binding to the N-terminal dimerization domain of H-NS and modifying its selectivity. Hha-like proteins and the H-NS N-terminal domain are unusually rich in charged residues, and their interaction is mostly electrostatic-driven but, nonetheless, highly selective. The NMR-based structural model of the complex between Hha/YmoA and the H-NS N-terminal dimerization domain reveals that the origin of the selectivity is the formation of a three-protein charge zipper with interdigitated complementary charged residues from Hha and the two units of the H-NS dimer. The free form of YmoA shows collective microsecond-millisecond dynamics that can by measured by NMR relaxation dispersion experiments and shows a linear dependence with the salt concentration. The number of residues sensing the collective dynamics and the population of the minor form increased in the presence of H-NS. Additionally, a single residue mutation in YmoA (D43N) abolished H-NS binding and the dynamics of the apo-form, suggesting the dynamics and binding are functionally related.
Keywords: charge zipper complexes; electrostatics; gene silencing; horizontal gene transfer; nuclear magnetic resonance (NMR); nucleoid-associated proteins; protein dynamic; protein-protein interaction.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- WHO (2014) Antimicrobial Resistance. Global Report on Surveillance, World Health Organization Press
-
- Ochman H., Lawrence J. G., Groisman E. A. (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 - PubMed
-
- Davies J. (1994) Inactivation of antibiotics and the dissemination of resistance genes. Science 264, 375–382 - PubMed
-
- Nieto J. M., Carmona M., Bolland S., Jubete Y., de la Cruz F., Juárez A. (1991) The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5, 1285–1293 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

