Linear ubiquitination in immunity
- PMID: 26085216
- PMCID: PMC4737190
- DOI: 10.1111/imr.12309
Linear ubiquitination in immunity
Abstract
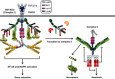
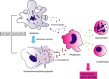
Linear ubiquitination is a post-translational protein modification recently discovered to be crucial for innate and adaptive immune signaling. The function of linear ubiquitin chains is regulated at multiple levels: generation, recognition, and removal. These chains are generated by the linear ubiquitin chain assembly complex (LUBAC), the only known ubiquitin E3 capable of forming the linear ubiquitin linkage de novo. LUBAC is not only relevant for activation of nuclear factor-κB (NF-κB) and mitogen-activated protein kinases (MAPKs) in various signaling pathways, but importantly, it also regulates cell death downstream of immune receptors capable of inducing this response. Recognition of the linear ubiquitin linkage is specifically mediated by certain ubiquitin receptors, which is crucial for translation into the intended signaling outputs. LUBAC deficiency results in attenuated gene activation and increased cell death, causing pathologic conditions in both, mice, and humans. Removal of ubiquitin chains is mediated by deubiquitinases (DUBs). Two of them, OTULIN and CYLD, are constitutively associated with LUBAC. Here, we review the current knowledge on linear ubiquitination in immune signaling pathways and the biochemical mechanisms as to how linear polyubiquitin exerts its functions distinctly from those of other ubiquitin linkage types.
Keywords: cell death; deubiquitinases; inflammation; linear ubiquitination; signaling pathways.
© 2015 The Authors. Immunological Reviews Published by John Wiley & Sons Ltd.
Figures
References
-
- Kulathu Y, Komander D. Atypical ubiquitylation – the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 2012;13:508–523. - PubMed
-
- Glickman MH, Ciechanover A. The ubiquitin‐proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002;82:373–428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

